Tuning Polyacrylate Composition to Recognize and Modulate Fluorescent Proteins
Authors:
Darwin C. Gomez, Swarnadeep Seth, Ronnie Mondal, Stephen J. Koehler, Jared G. Baker, Charles Plate, Ian C. Anderson, Mikayla R. Smith, Joey Gloriod, Morgan Gunter, Valerie V. Welborn,* Sanket A. Deshmukh, and C. Adrian Figg
Affiliation:
D. C. Gomez, R. Mondal, S. J. Koehler, J. G. Baker, I. C. Anderson, M. R. Smith, J. Gloriod, M. Gunter, V. V. Welborn, C. A. Figg Macromolecules Innovation Institute and Department of Chemistry, Virginia Tech, Blacksburg, Virginia 24061, USA
D. C. Gomez, S. J. Koehler, J. G. Baker, I. C. Anderson, M. R. Smith, J. Gloriod, M. Gunter, C. A. Figg Virginia Tech Center for Drug Discovery, Virginia Tech, Blacksburg,Virginia 24061, USA
D. C. Gomez Department of Chemistry, Eastern Visayas State University, Tacloban City 6500, Philippines
S. Seth, C. Plate, S. A. Deshmukh Department of Chemical Engineering, Virginia Tech, Blacksburg, Virginia 24061, USA
Description:
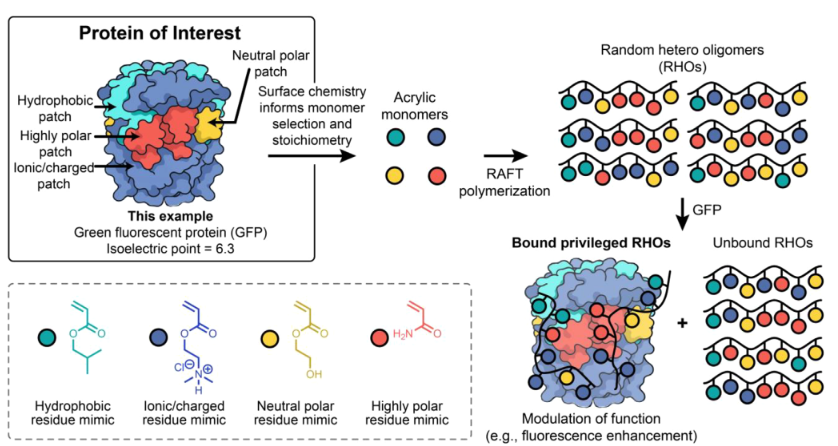
Molecular definition is usually regarded as a prerequisite to achieve protein recognition and functionalmodulation, particularly for macromolecular interactions. Herein, we report that polymers with specific combinations ofmonomers arranged into random sequences [random hetero oligomers (RHOs)] can selectively bind to a model protein.Using green fluorescent protein (GFP) as a target, polyacrylates were developed that bound with nanomolar affinity andenhanced fluorescence by >100%. Purification of the polymerization product revealed subpopulations of compositionswith distinct affinities and selectivity for GFP over a competing protein. Experimental and computational binding analysesconfirmed that there are distinct RHO–GFP interactions, which are influenced by RHO chemical composition. Thesefindings show that sequence-defined structures are not a prerequisite for selective protein recognition. Synthetic polymerscan instead serve as scalable, tunable platforms for molecular recognition—representing a significant leap towards next-generation sensing, therapeutic, responsive, and catalytic materials in domains previously dominated by biologics orcomplex peptide scaffolds.
Publications:
- Darwin C. Gomez, Swarnadeep Seth, Ronnie Mondal, Stephen J. Koehler, Jared G. Baker, Charles Plate, Ian C. Anderson, Mikayla R. Smith, Joey Gloriod, Morgan Gunter, Valerie V. Welborn,* Sanket A. Deshmukh, and C. Adrian Figg; Tuning Polyacrylate Composition to Recognize and Modulate Fluorescent Proteins; Angewandte Chemie International Edition, 2026
Tags:
Polymers ProteinRelated Chronicles:
No related chronicles available
Files:
| File Name | File Description | File Type | File Size | File URL |
|---|---|---|---|---|
| Supplementary Information | Experimental and simulation supporting information. | docx | 12.23 MB | Login to download |
| Simulation Movies | Movies on metadynamics simulations of oligomer-protein free energy landscapes. | zip | 43.51 MB | Login to download |