Complex Carbohydrate Magnetic Resonance Database (CCMRD)
Carbohydrates are crucial for various life activities in living organisms. The Complex Carbohydrates Magnetic Resonance Database (CCMRD), a solid-state NMR database for complex carbohydrates developed at Michigan State University. The Solid-state NMR spectroscopy reveals the molecular structure and dynamics of insoluble complex carbohydrates.
| Database ID | Trival Name | Linear Code | Compound Class | Taxonomy Domain | Species | Residue | SNFG | Chemical Structure | Chemical Shifts (ppm) | Spectrometer (MHz) | Temperature (K) | pH | NMR Reference Compound | Sample Treatment | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ccmrd_678 | cellulose | Glcp | Polysaccharide | plant | Eucalyptus grandis | b-D-Glcp |
|
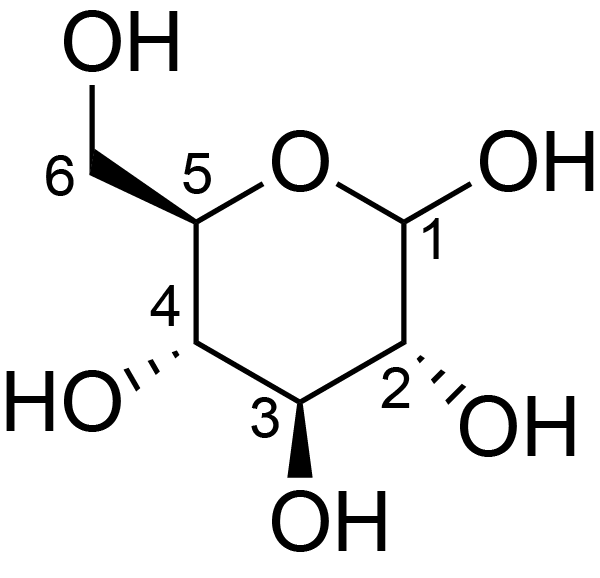
|
C5: 73.7, C6: 61.2 | 600/400 | 298.0 | None | TMS | whole cell | Alex Kirui et al., Carbohydrate-aromatic interface and molecular architecture of lignocellulose, NATURE COMMUNICATIONS, 2022 |
| ccmrd_679 | Xylan | Xylp | Polysaccharide | plant | Eucalyptus grandis | ?-?-Xylp |
|
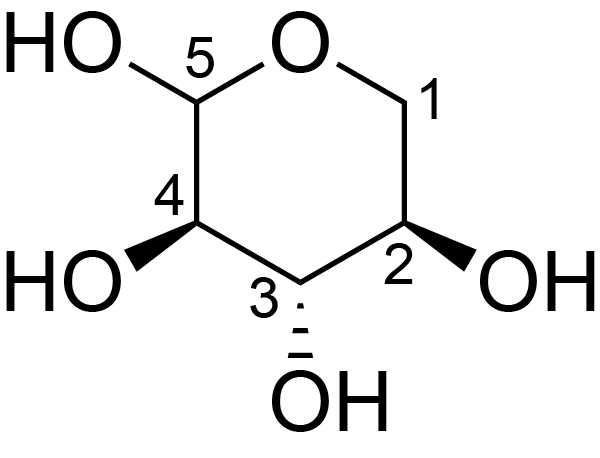
|
C1: 104.8, C2: 72.2, C3: 74.0, C4: 82.2, C5: 64.0 | 600/400 | 298.0 | None | TMS | whole cell | Alex Kirui et al., Carbohydrate-aromatic interface and molecular architecture of lignocellulose, NATURE COMMUNICATIONS, 2022 |
| ccmrd_680 | Xylan | Xylp | Polysaccharide | plant | Eucalyptus grandis | ?-?-Xylp |
|
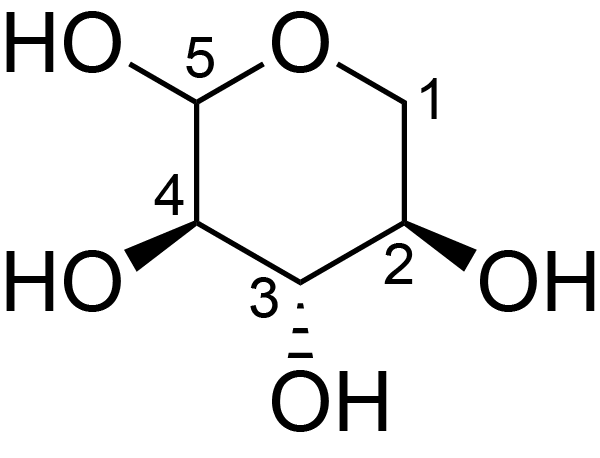
|
C1: 102.5, C2: 73.4, C3: 74.1, C4: 77.4, C5: 63.3 | 600/400 | 298.0 | None | TMS | whole cell | Alex Kirui et al., Carbohydrate-aromatic interface and molecular architecture of lignocellulose, NATURE COMMUNICATIONS, 2022 |
| ccmrd_681 | Xylan | Xylp | Polysaccharide | plant | Eucalyptus grandis | ?-?-Xylp |
|
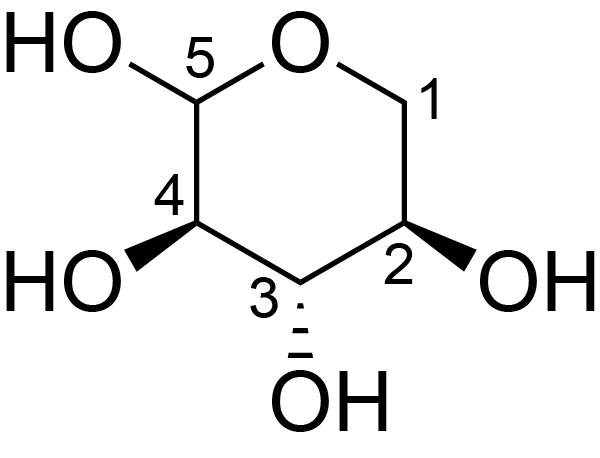
|
C3: 74.2, C4: 78.5, C5: 64.1 | 600/400 | 298.0 | None | TMS | whole cell | Alex Kirui et al., Carbohydrate-aromatic interface and molecular architecture of lignocellulose, NATURE COMMUNICATIONS, 2022 |
| ccmrd_682 | Acetyl-methyl groups | Ac | AcMe Groups | plant | Eucalyptus grandis | ?-?-Ac |
|
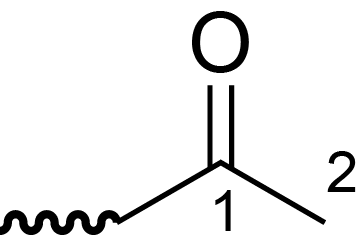
|
C1: 174.0, C2: 21.0 | 600/400 | 298.0 | None | TMS | whole cell | Alex Kirui et al., Carbohydrate-aromatic interface and molecular architecture of lignocellulose, NATURE COMMUNICATIONS, 2022 |
| ccmrd_683 | cellulose | Glcp | Polysaccharide | plant | Eucalyptus grandis | b-D-Glcp |
|
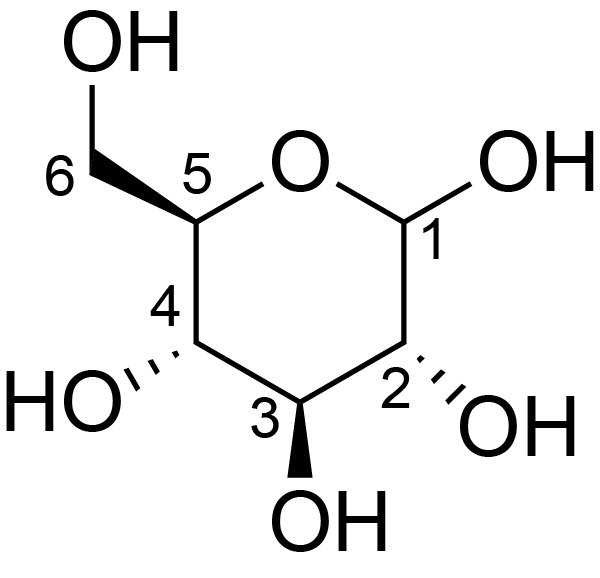
|
C1: 104.9, C2: 72.1, C3: 75.2, C4: 88.7, C5: 72.3, C6: 65.0 | 600/400 | 298.0 | None | TMS | whole cell | Alex Kirui et al., Carbohydrate-aromatic interface and molecular architecture of lignocellulose, NATURE COMMUNICATIONS, 2022 |
| ccmrd_684 | cellulose | Glcp | Polysaccharide | plant | Eucalyptus grandis | ?-?-Glcp |
|
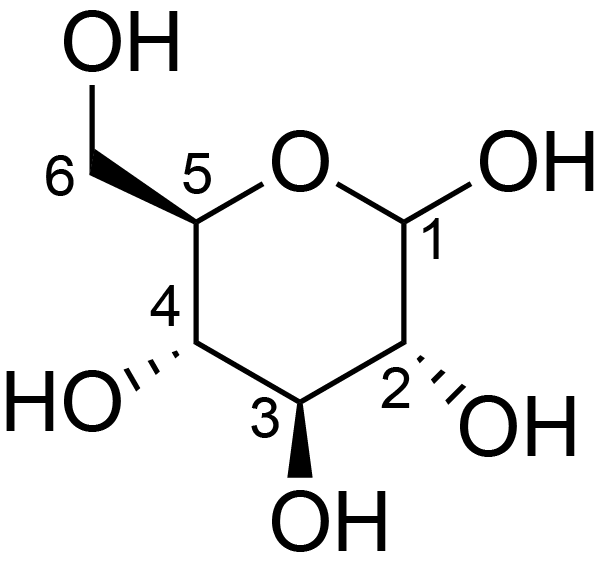
|
C1: 104.9, C2: 72.1, C3: 74.1, C4: 87.9, C5: 71.1, C6: 65.7 | 600/400 | 298.0 | None | TMS | whole cell | Alex Kirui et al., Carbohydrate-aromatic interface and molecular architecture of lignocellulose, NATURE COMMUNICATIONS, 2022 |
| ccmrd_685 | cellulose | Glcp | Polysaccharide | plant | Eucalyptus grandis | b-D-Glcp |
|
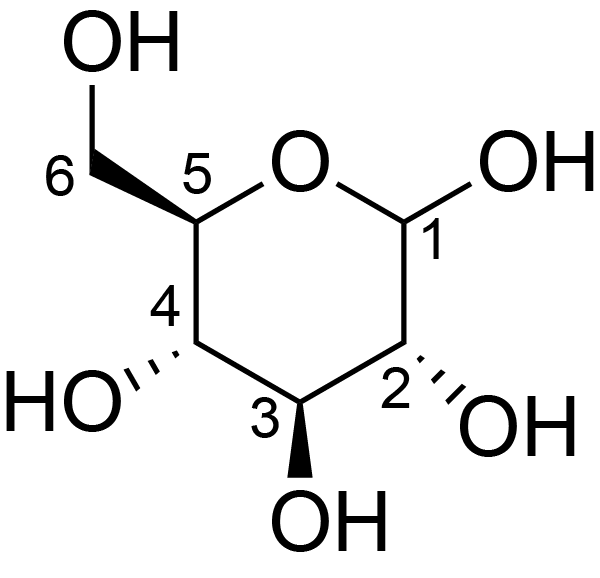
|
C1: 104.9, C2: 72.1, C3: 74.6, C4: 84.4, C5: 74.6, C6: 62.3 | 600/400 | 298.0 | None | TMS | whole cell | Alex Kirui et al., Carbohydrate-aromatic interface and molecular architecture of lignocellulose, NATURE COMMUNICATIONS, 2022 |
| ccmrd_686 | cellulose | Glcp | Polysaccharide | plant | Eucalyptus grandis | b-D-Glcp |
|
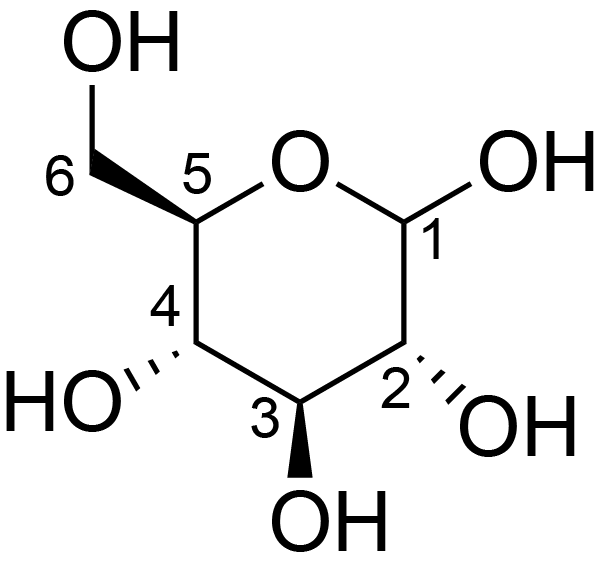
|
C1: 104.9, C2: 72.1, C3: 73.6, C4: 83.5, C5: 73.6, C6: 61.4 | 600/400 | 298.0 | None | TMS | whole cell | Alex Kirui et al., Carbohydrate-aromatic interface and molecular architecture of lignocellulose, NATURE COMMUNICATIONS, 2022 |
| ccmrd_687 | cellulose | Glcp | Polysaccharide | plant | Populus × canadensis | b-D-Glcp |
|
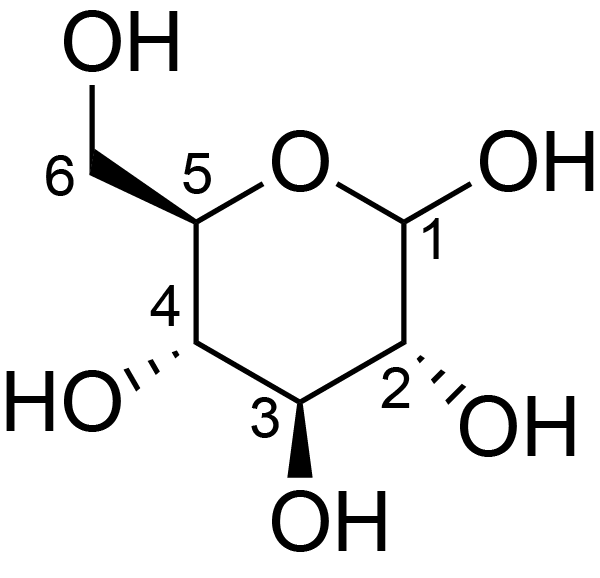
|
C3: 75.5, C4: 88.9, C5: 72.4, C6: 64.9 | 600/400 | 298.0 | None | TMS | whole cell | Alex Kirui et al., Carbohydrate-aromatic interface and molecular architecture of lignocellulose, NATURE COMMUNICATIONS, 2022 |