Complex Carbohydrate Magnetic Resonance Database (CCMRD)
Carbohydrates are crucial for various life activities in living organisms. The Complex Carbohydrates Magnetic Resonance Database (CCMRD), a solid-state NMR database for complex carbohydrates developed at Michigan State University. The Solid-state NMR spectroscopy reveals the molecular structure and dynamics of insoluble complex carbohydrates.
| Database ID | Trival Name | Linear Code | Compound Class | Taxonomy Domain | Species | Residue | SNFG | Chemical Structure | Chemical Shifts (ppm) | Spectrometer (MHz) | Temperature (K) | pH | NMR Reference Compound | Sample Treatment | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ccmrd_466 | Chitin | GlcpNAc | Polysaccharide | fungi | Aspergillus nidulans | ?-D-GlcpNAc |
|
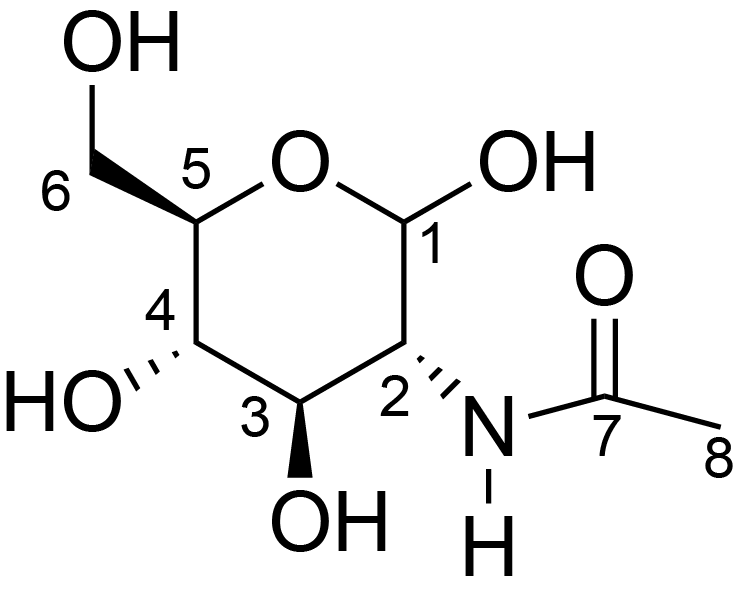
|
C1: 102.6, C2: 54.5, C3: 73.6, C4: 83.1, C5: 75.4, C6: 60.7, C7: 173.4, C8: 22.3 | 800/850 | 298.0 | None | TMS | whole cell, cultured with 0.5M NaCl | Liyanage et al., Structural, Frontiers, 2021 |
| ccmrd_467 | Chitin | GlcpNAc | Polysaccharide | fungi | Aspergillus sydowii | ?-D-GlcpNAc |
|
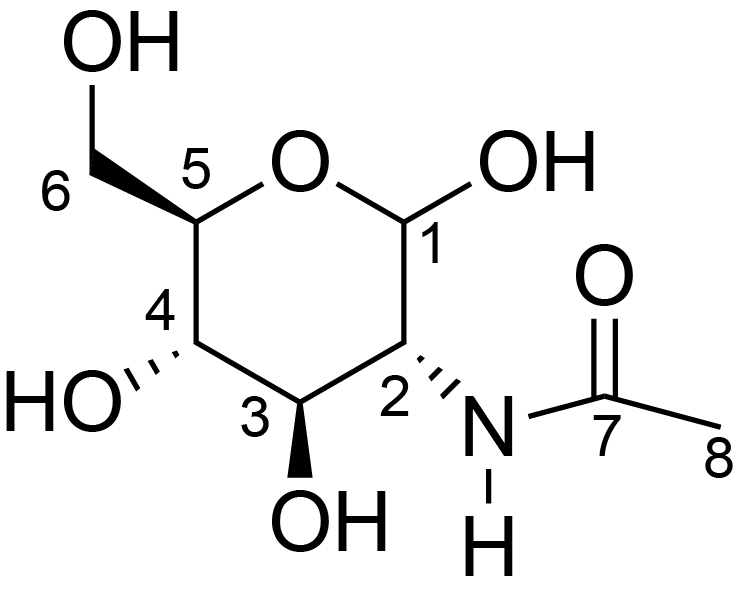
|
C1: 103.3, C2: 55.5, C3: 72.9, C4: 84.5, C5: 76.2, C6: 60.7, C7: 174.6, C8: 22.7 | 800/850 | 298.0 | None | TMS | whole cell, cultured with 0.5M NaCl | Liyanage et al., Structural, Frontiers, 2021 |
| ccmrd_468 | chitosan | GlcpN | Polysaccharide | fungi | Aspergillus sydowii | ?-D-GlcpN |
|
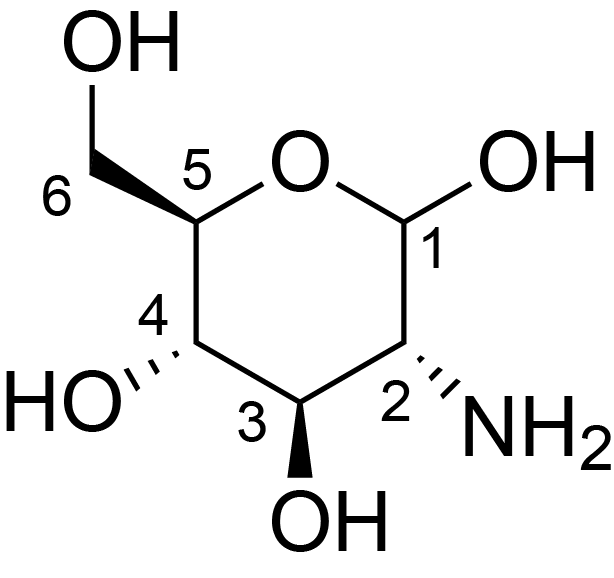
|
C1: 102.2, C2: 55.6, C3: 74.5, C4: 80.4, C5: 74.9, C6: 60.7 | 800/850 | 298.0 | None | TMS | whole cell, cultured with 0.5M NaCl | Liyanage et al., Structural, Frontiers, 2021 |
| ccmrd_469 | Chitin | GlcpNAc | Polysaccharide | fungi | Aspergillus sydowii | ?-D-GlcpNAc |
|
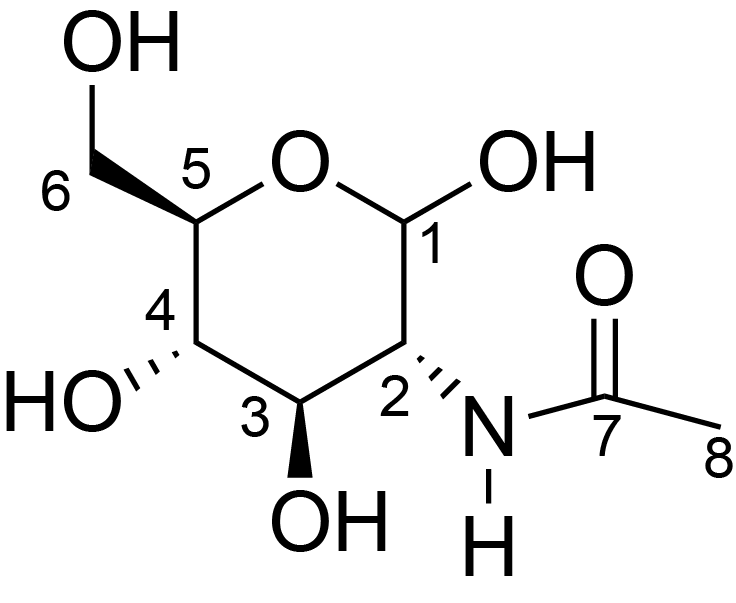
|
C1: 103.5, C2: 55.2, C3: 73.4, C4: 84.4, C5: 75.9, C6: 60.0, C7: 173.4, C8: 22.7 | 800/850 | 298.0 | None | TMS | whole cell, cultured with 0.5M NaCl | Liyanage et al., Structural, Frontiers, 2021 |
| ccmrd_470 | Chitin | GlcpNAc | Polysaccharide | fungi | Aspergillus sydowii | ?-D-GlcpNAc |
|
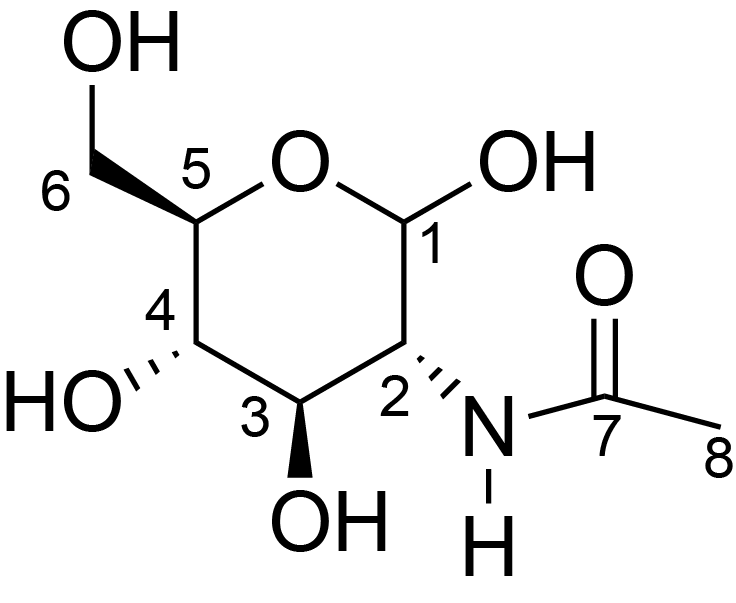
|
C1: 103.5, C2: 55.4, C3: 73.3, C4: 83.7, C5: 75.9, C6: 60.7, C7: 173.4, C8: 22.7 | 800/850 | 298.0 | None | TMS | whole cell, cultured with 0.5M NaCl | Liyanage et al., Structural, Frontiers, 2021 |
| ccmrd_471 | Chitin | GlcpNAc | Polysaccharide | fungi | Aspergillus sydowii | ?-D-GlcpNAc |
|
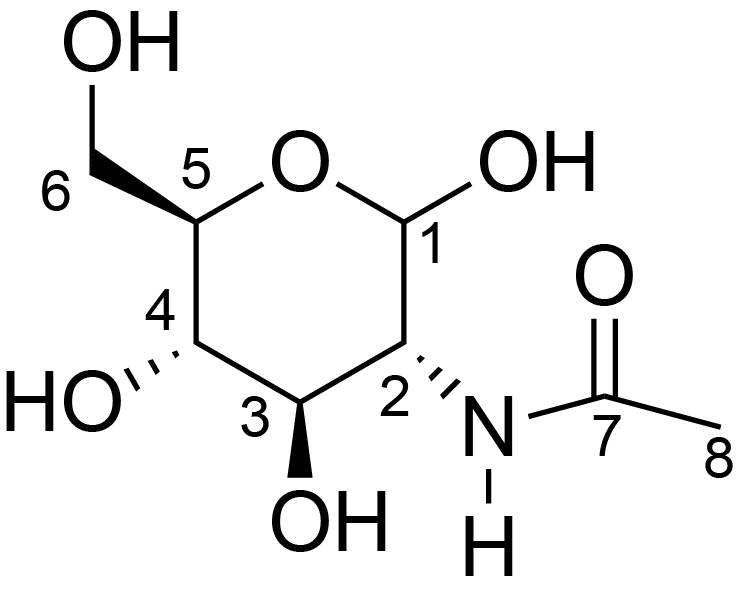
|
C1: 103.6, C2: 55.0, C3: 73.2, C4: 83.4, C5: 75.5, C6: 60.5, C7: 174.1, C8: 22.4 | 800/850 | 298.0 | None | TMS | whole cell, cultured with 0.5M NaCl | Liyanage et al., Structural, Frontiers, 2021 |
| ccmrd_472 | Chitin | GlcpNAc | Polysaccharide | fungi | Aspergillus sydowii | ?-D-GlcpNAc |
|
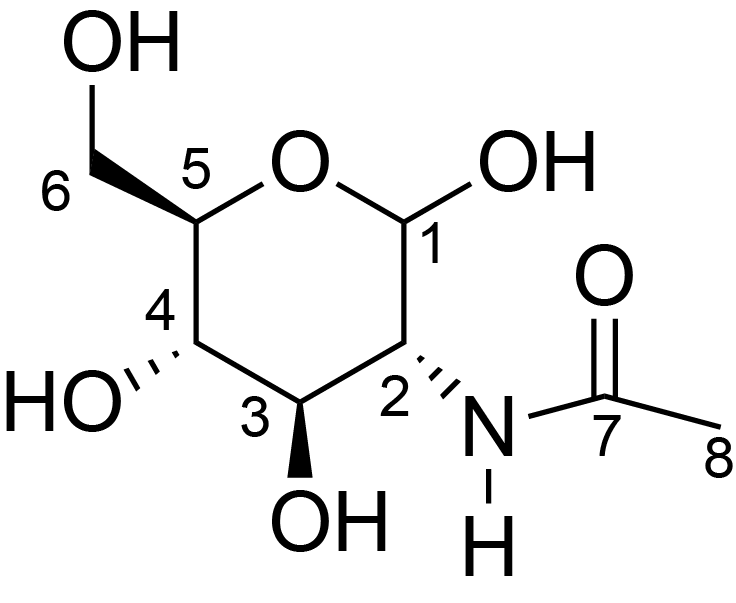
|
C1: 103.2, C2: 54.8, C3: 73.5, C4: 82.5, C5: 75.2, C6: 60.9, C7: 175.1, C8: 22.4 | 800/850 | 298.0 | None | TMS | whole cell, cultured with 0.5M NaCl | Liyanage et al., Structural, Frontiers, 2021 |
| ccmrd_473 | chitosan | GlcpN | Polysaccharide | fungi | Aspergillus sydowii | ?-D-GlcpN |
|
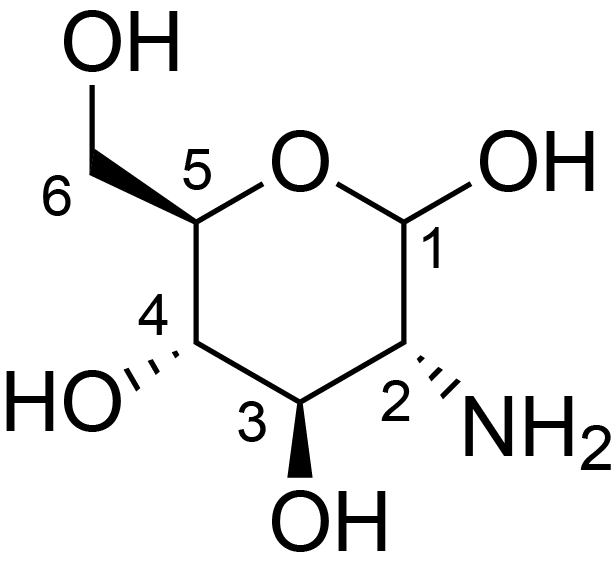
|
C1: 101.9, C2: 55.7, C3: 72.9, C4: 80.0, C5: 74.3, C6: 60.5 | 800/850 | 298.0 | None | TMS | whole cell, cultured with 0.5M NaCl | Liyanage et al., Structural, Frontiers, 2021 |
| ccmrd_474 | chitosan | GlcpN | Polysaccharide | fungi | Aspergillus sydowii | ?-D-GlcpN |
|
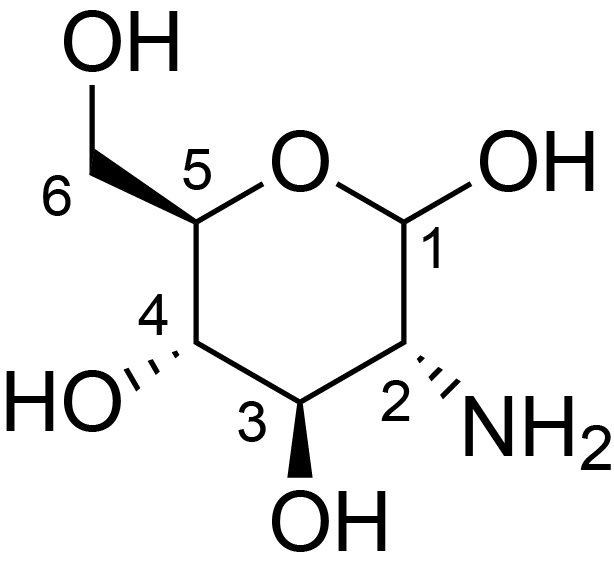
|
C1: 102.9, C2: 55.7, C3: 72.5, C4: 79.7, C5: 75.3, C6: 60.9 | 800/850 | 298.0 | None | TMS | whole cell, cultured with 0.5M NaCl | Liyanage et al., Structural, Frontiers, 2021 |
| ccmrd_475 | chitosan | GlcpN | Polysaccharide | fungi | Aspergillus sydowii | ?-D-GlcpN |
|
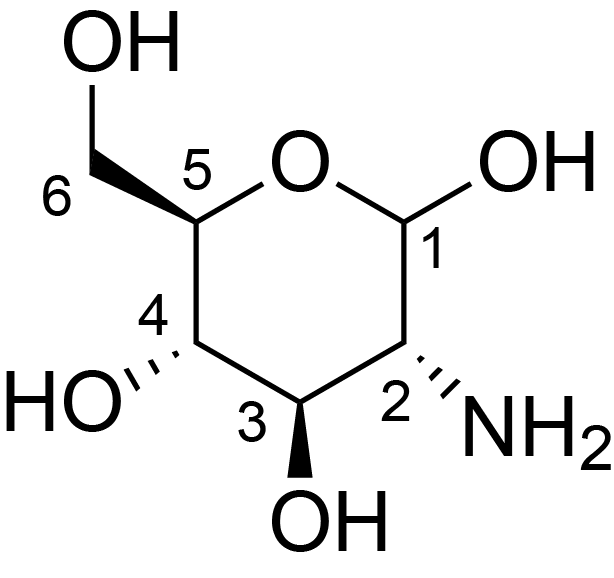
|
C1: 101.4, C2: 55.5, C3: 73.5, C4: 79.1, C5: 75.3, C6: 61.0 | 800/850 | 298.0 | None | TMS | whole cell, cultured with 0.5M NaCl | Liyanage et al., Structural, Frontiers, 2021 |