Complex Carbohydrate Magnetic Resonance Database (CCMRD)
Carbohydrates are crucial for various life activities in living organisms. The Complex Carbohydrates Magnetic Resonance Database (CCMRD), a solid-state NMR database for complex carbohydrates developed at Michigan State University. The Solid-state NMR spectroscopy reveals the molecular structure and dynamics of insoluble complex carbohydrates.
| Database ID | Trival Name | Linear Code | Compound Class | Taxonomy Domain | Species | Residue | SNFG | Chemical Structure | Chemical Shifts (ppm) | Spectrometer (MHz) | Temperature (K) | pH | NMR Reference Compound | Sample Treatment | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ccmrd_446 | teichoic acid | GlcpNAcMurpNAcManpNAcRibitolAATGalp | Peptidoglycan | bacteria | Streptococcus pneumoniae | a-?-GalpNAc |
|

|
C1: 94.1, C2: 50.2, C3: 67.6, C4: 77.3, C5: 71.3, C6: 64.3, C7: 175.4, C8: 23.0 | 400 and 600 | 285.0 | None | ND | Being resuspended in HEPES buffer pH 7.5 | Thomas Kern et al., Dynamics Characterization of Fully Hydrated Bacterial Cell Walls by Solid-State NMR: Evidence for Cooperative Binding of Metal Ions, Journal of the American Chemical Society, 2010 |
| ccmrd_447 | teichoic acid | GlcpNAcMurpNAcManpNAcRibitolAATGalp | Peptidoglycan | bacteria | Streptococcus pneumoniae | b-?-Glcp |
|
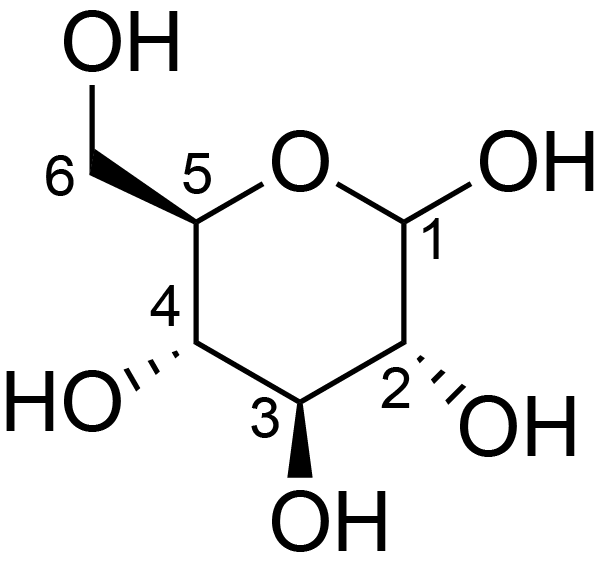
|
C1: 104.8, C2: 73.6, C3: 76.3, C4: 69.6, C5: 75.1, C6: 64.9 | 400 and 600 | 285.0 | None | ND | Being resuspended in HEPES buffer pH 7.5 | Thomas Kern et al., Dynamics Characterization of Fully Hydrated Bacterial Cell Walls by Solid-State NMR: Evidence for Cooperative Binding of Metal Ions, Journal of the American Chemical Society, 2010 |
| ccmrd_448 | teichoic acid | GlcpNAcMurpNAcManpNAcRibitolAATGalp | Peptidoglycan | bacteria | Streptococcus pneumoniae | ?-?-AAt-Galp |
|

|
C1: 98.9, C2: 49.0, C3: 75.8, C4: 55.5, C5: 64.1, C6: 16.3 | 400 and 600 | 285.0 | None | ND | Being resuspended in HEPES buffer pH 7.5 | Thomas Kern et al., Dynamics Characterization of Fully Hydrated Bacterial Cell Walls by Solid-State NMR: Evidence for Cooperative Binding of Metal Ions, Journal of the American Chemical Society, 2010 |
| ccmrd_449 | Rhamnogalacturonan I | (1-4)αDGalpA(1-2)αLRhap | Polysaccharide | plant | Arabidopsis thaliana | a-L-Rhap |
|
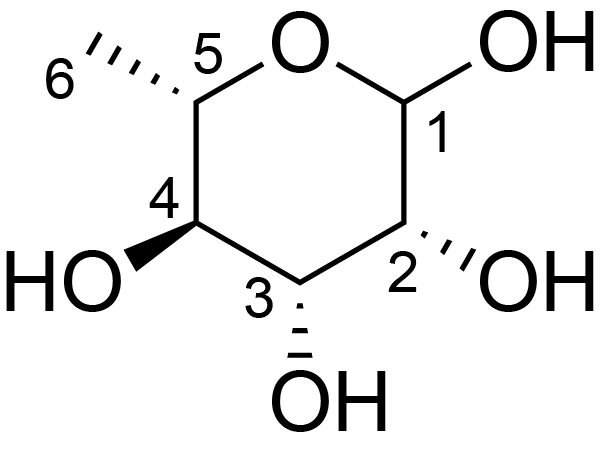
|
H5: 3.9, H6: 1.3 | 400, 600, and 800 | 298.0 | None | TMS | Removing lipid membranes, cytoplasmic proteins, small molecules, and starch. And being back-exchanged in D2O to reduce the water 1H signal intensities. | Pyae Phyo and Mei Hong et al., Fast MAS 1H-13C correlation NMR for structural investigations of plant cell walls, Journal of Biomolecular NMR, 2019 |
| ccmrd_450 | Rhamnogalacturonan I | (1-4)αDGalpA(1-2)αLRhap | Polysaccharide | plant | Arabidopsis thaliana | a-L-Rhap |
|
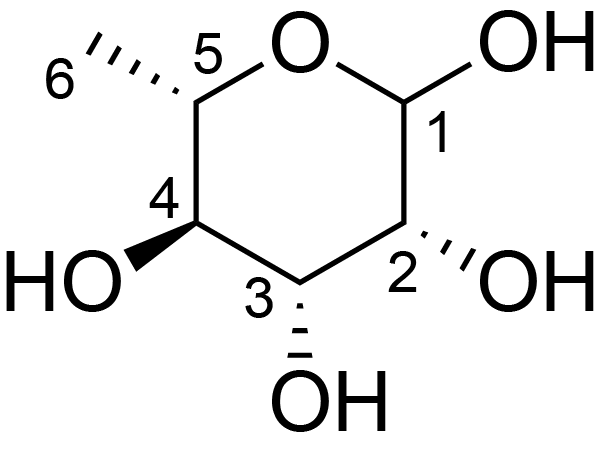
|
H1: 5.1, H2: 4.3, H4: 3.6, H5: 4.0, H6: 1.2 | 400, 600, and 800 | 298.0 | None | TMS | Removing lipid membranes, cytoplasmic proteins, small molecules, and starch. And being back-exchanged in D2O to reduce the water 1H signal intensities. | Pyae Phyo and Mei Hong et al., Fast MAS 1H-13C correlation NMR for structural investigations of plant cell walls, Journal of Biomolecular NMR, 2019 |
| ccmrd_451 | Rhamnogalacturonan I | (1-4)αDGalpA(1-2)αLRhap | Polysaccharide | plant | Arabidopsis thaliana | a-L-Rhap |
|
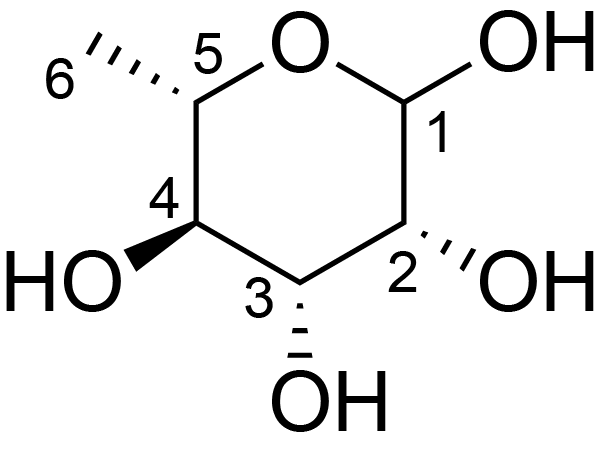
|
H1: 5.1, H2: 4.4, H4: 3.6, H5: 4.0, H6: 1.2 | 400, 600, and 800 | 298.0 | None | TMS | Removing lipid membranes, cytoplasmic proteins, small molecules, and starch. And being back-exchanged in D2O to reduce the water 1H signal intensities. | Pyae Phyo and Mei Hong et al., Fast MAS 1H-13C correlation NMR for structural investigations of plant cell walls, Journal of Biomolecular NMR, 2019 |
| ccmrd_452 | cellulose | Glcp | Polysaccharide | plant | Picea abies (Norway spruce) | b-D-Glcp |
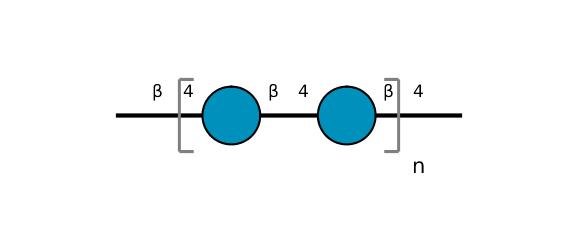
|
|
C1: 104.3, C4: 88.3, C5: 71.7, C6: 66.0 | 850 | 298.0 | None | TMS | frozen in liquid nitrogen and ground to produce a fine powder | Oliver M. Terrett et al., Molecular architecture of softwood revealed by solid-state NMR, Nature Communications, 2019 |
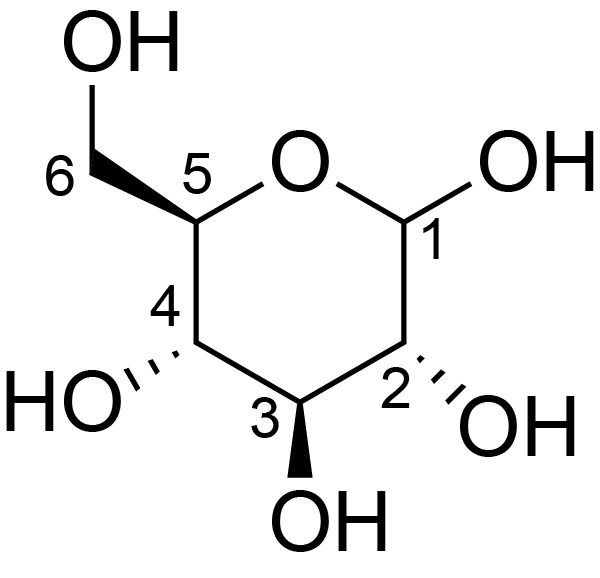
| ccmrd_453 | cellulose II oligomer crystals | Glcp | oligosachaaride | other | cellulose II oligomer crystals | ?-?-Glcp |
|
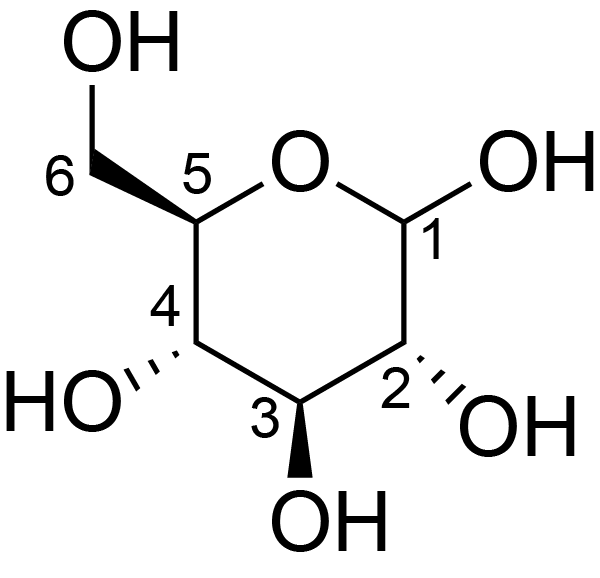
|
C1: 92.7, C2: 73.0, C3: 75.4, C4: 88.3, C5: 75.4, C6: 63.2 | 700 | 298.0 | None | TMS | reducing ends | Yusuke Kita . Ryosuke Kusumi . Tsunehisa Kimura . Motomitsu Kitaoka . Yusuke Nishiyama . Masahisa Wada et al., Surface structural analysis of selectively 13C-labeled cellulose II by solid-state NMR spectroscopy, Cellulose, 2020 |
| ccmrd_454 | cellulose II oligomer crystals | Glcp | oligosachaaride | other | cellulose II oligomer crystals | ?-?-Glcp |
|
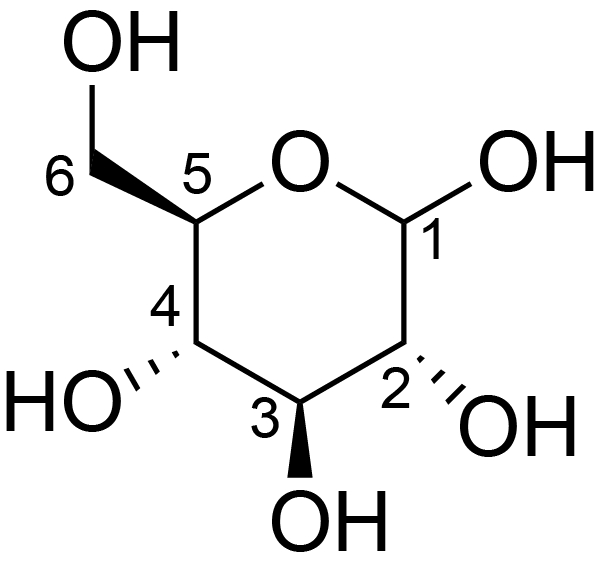
|
C1: 96.5, C2: 76.1, C3: 77.2, C4: 88.7, C5: 73.5, C6: 64.3 | 700 | 298.0 | None | TMS | reducing ends | Yusuke Kita . Ryosuke Kusumi . Tsunehisa Kimura . Motomitsu Kitaoka . Yusuke Nishiyama . Masahisa Wada et al., Surface structural analysis of selectively 13C-labeled cellulose II by solid-state NMR spectroscopy, Cellulose, 2020 |
| ccmrd_455 | Chitin | GlcpNAc | Polysaccharide | fungi | Aspergillus fumigatus | ?-D-GlcpNAc |
|
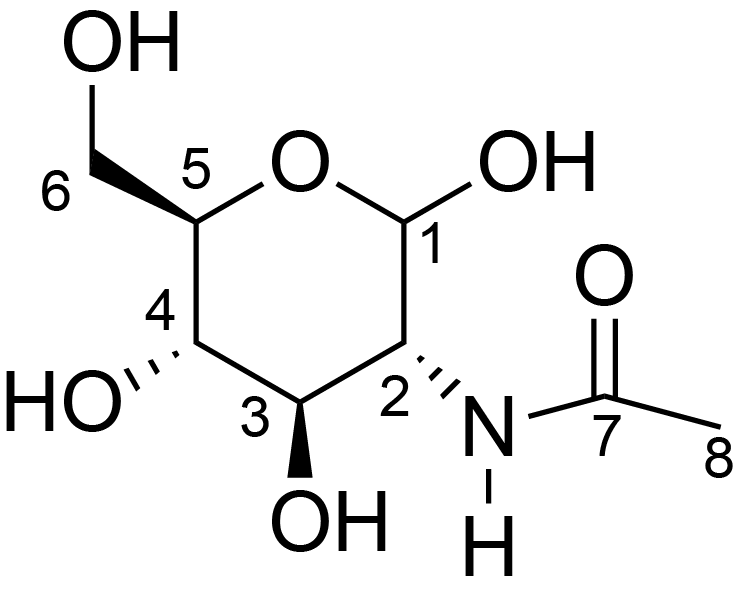
|
C1: 104.3, C2: 55.7, C3: 73.2, C4: 83.9, C5: 75.2, C6: 60.7, C7: 173.9, C8: 23.3 | 800/850 | 298.0 | None | TMS | whole cell, cultured with 0.5M NaCl | Liyanage et al., Structural, Frontiers, 2021 |