Complex Carbohydrate Magnetic Resonance Database (CCMRD)
Carbohydrates are crucial for various life activities in living organisms. The Complex Carbohydrates Magnetic Resonance Database (CCMRD), a solid-state NMR database for complex carbohydrates developed at Michigan State University. The Solid-state NMR spectroscopy reveals the molecular structure and dynamics of insoluble complex carbohydrates.
| Database ID | Trival Name | Linear Code | Compound Class | Taxonomy Domain | Species | Residue | SNFG | Chemical Structure | Chemical Shifts (ppm) | Spectrometer (MHz) | Temperature (K) | pH | NMR Reference Compound | Sample Treatment | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ccmrd_416 | Galactoglucomannan | bDGlcp(1-4)[aDGalp(1-6)]bDManp(1-4)bDManp(1-4)bDGlcp(1-4)bDGlcp(1-4)[aDGalp(1-6)]bDManp(1-4)bDGlcp(1-4) | Polysaccharide | plant | Picea abies (Norway spruce) | b-D-Manp |
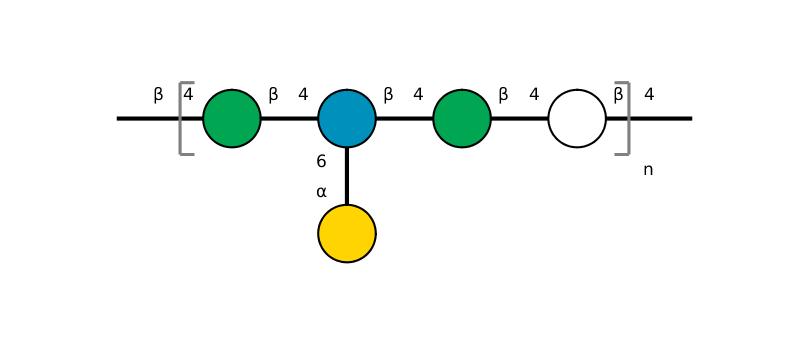
|
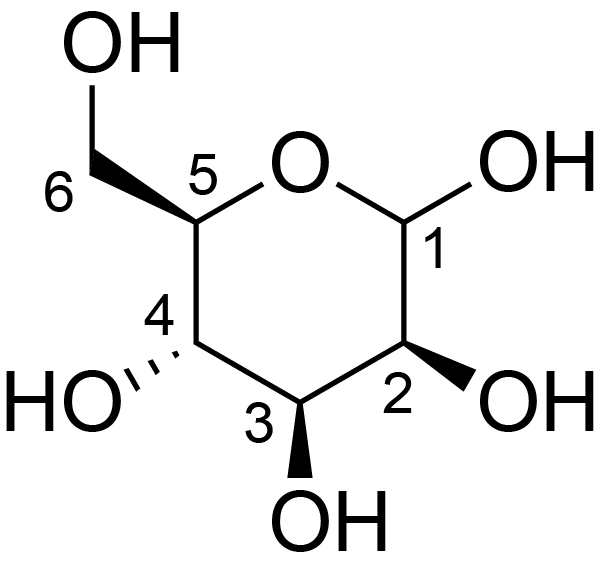
|
C1: 101.9, C2: 72.0, C4: 80.4, C5: 75.8, C6: 61.6 | 850 | 298.0 | None | TMS | frozen in liquid nitrogen and ground to produce a fine powder | Oliver M. Terrett et al., Molecular architecture of softwood revealed by solid-state NMR, Nature Communications, 2019 |
| ccmrd_417 | Acetylated Galactoglucomannan | bDGlcp(1-4)[aDGalp(1-6)]bDManp(1-4)bDManp(1-4)bDGlcp(1-4)bDGlcp(1-4)[aDGalp(1-6)]bDManp(1-4)bDGlcp(1-4) | Polysaccharide | plant | Picea abies (Norway spruce) | b-D-ManpOAc |
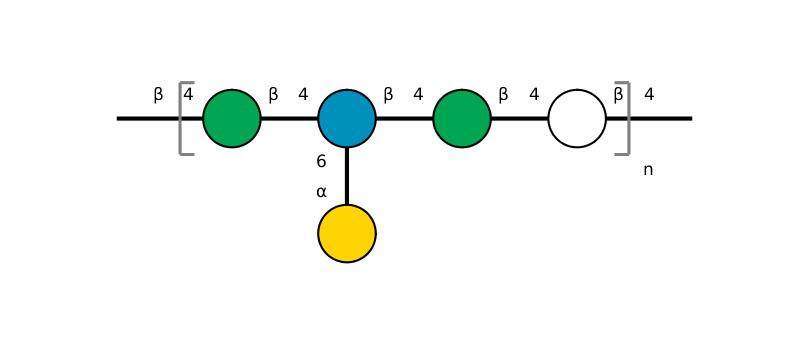
|

|
C1: 100.9, C2: 71.9, C3: 75.9, C4: 80.4, C5: 75.8, C6: 61.6 | 850 | 298.0 | None | TMS | frozen in liquid nitrogen and ground to produce a fine powder | Oliver M. Terrett et al., Molecular architecture of softwood revealed by solid-state NMR, Nature Communications, 2019 |
| ccmrd_418 | xylan or Arabinogalactan | Xylp or [bLAraf(1-3)bLAraf(1-6)]bDGalp(1-3) | Polysaccharide | plant | Picea abies (Norway spruce) | ?-?-Araf |
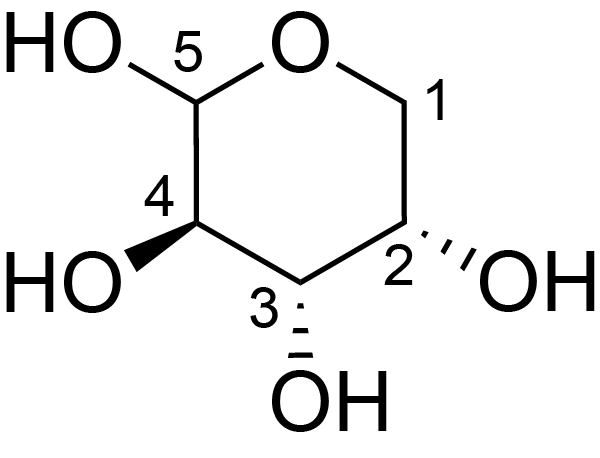
|
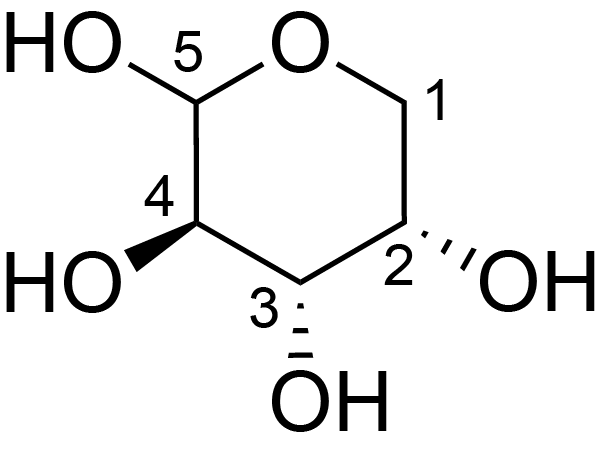
|
C1: 108.6, C2: 82.1, C3: 78.3, C4: 86.1, C5: 62.8 | 850 | 298.0 | None | TMS | frozen in liquid nitrogen and ground to produce a fine powder | Oliver M. Terrett et al., Molecular architecture of softwood revealed by solid-state NMR, Nature Communications, 2019 |
| ccmrd_419 | Rhamnogalacturonan I | (1-4)αDGalpA(1-2)αLRhap | Polysaccharide | plant | Arabidopsis thaliana | a-L-Araf |
|
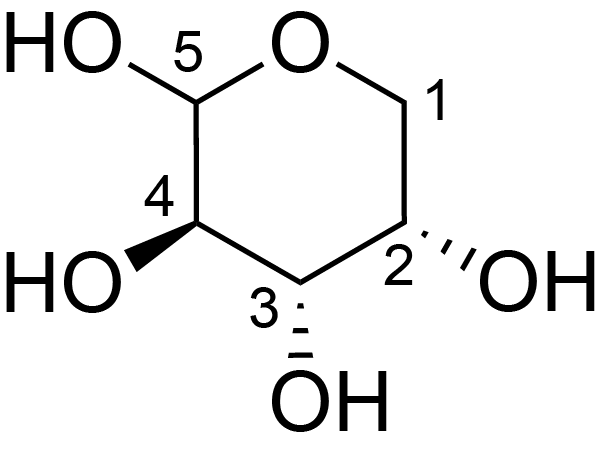
|
H1: 5.1, H2: 4.1 | 400, 600, and 800 | 298.0 | None | TMS | Removing lipid membranes, cytoplasmic proteins, small molecules, and starch. And being back-exchanged in D2O to reduce the water 1H signal intensities. | Pyae Phyo and Mei Hong et al., Fast MAS 1H-13C correlation NMR for structural investigations of plant cell walls, Journal of Biomolecular NMR, 2019 |
| ccmrd_420 | Rhamnogalacturonan I | (1-4)αDGalpA(1-2)αLRhap | Polysaccharide | plant | Arabidopsis thaliana | a-L-Araf |
|
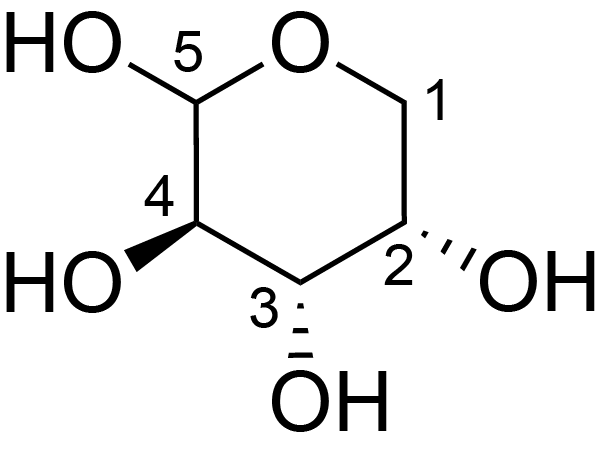
|
H1: 5.2, H2: 4.3 | 400, 600, and 800 | 298.0 | None | TMS | Removing lipid membranes, cytoplasmic proteins, small molecules, and starch. And being back-exchanged in D2O to reduce the water 1H signal intensities. | Pyae Phyo and Mei Hong et al., Fast MAS 1H-13C correlation NMR for structural investigations of plant cell walls, Journal of Biomolecular NMR, 2019 |
| ccmrd_421 | Rhamnogalacturonan I | (1-4)αDGalpA(1-2)αLRhap | Polysaccharide | plant | Arabidopsis thaliana | a-L-Araf |
|
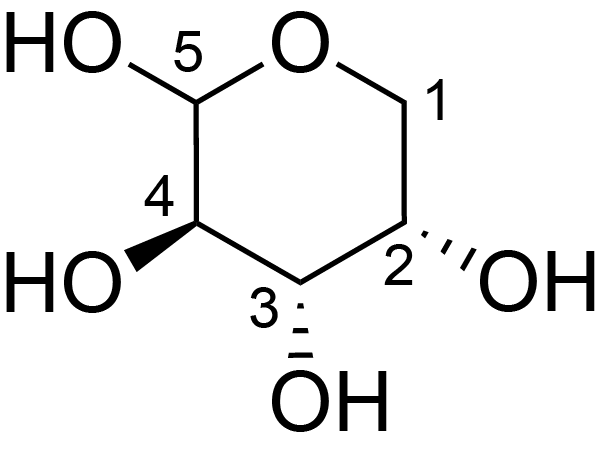
|
H1: 5.0, H2: 4.1, H3: 4.0, H4: 4.2, H5: 3.8 | 400, 600, and 800 | 298.0 | None | TMS | Removing lipid membranes, cytoplasmic proteins, small molecules, and starch. And being back-exchanged in D2O to reduce the water 1H signal intensities. | Pyae Phyo and Mei Hong et al., Fast MAS 1H-13C correlation NMR for structural investigations of plant cell walls, Journal of Biomolecular NMR, 2019 |
| ccmrd_422 | Rhamnogalacturonan I | (1-4)αDGalpA(1-2)αLRhap | Polysaccharide | plant | Arabidopsis thaliana | a-L-Araf |
|
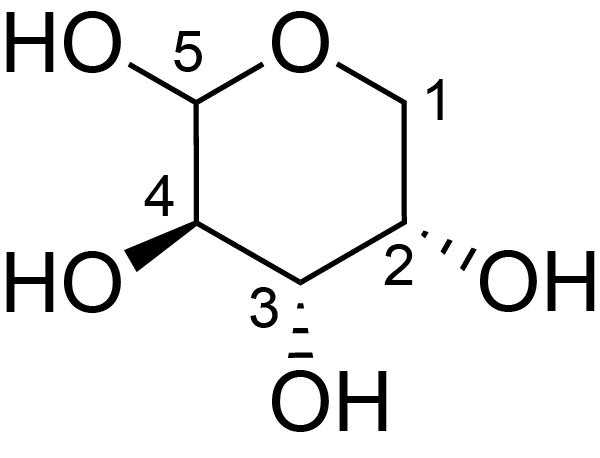
|
H1: 5.1, H2: 4.1, H3: 3.9, H4: 4.0, H5: 3.7 | 400, 600, and 800 | 298.0 | None | TMS | Removing lipid membranes, cytoplasmic proteins, small molecules, and starch. And being back-exchanged in D2O to reduce the water 1H signal intensities. | Pyae Phyo and Mei Hong et al., Fast MAS 1H-13C correlation NMR for structural investigations of plant cell walls, Journal of Biomolecular NMR, 2019 |
| ccmrd_423 | Rhamnogalacturonan I | (1-4)αDGalpA(1-2)αLRhap | Polysaccharide | plant | Arabidopsis thaliana | a-L-Araf |
|
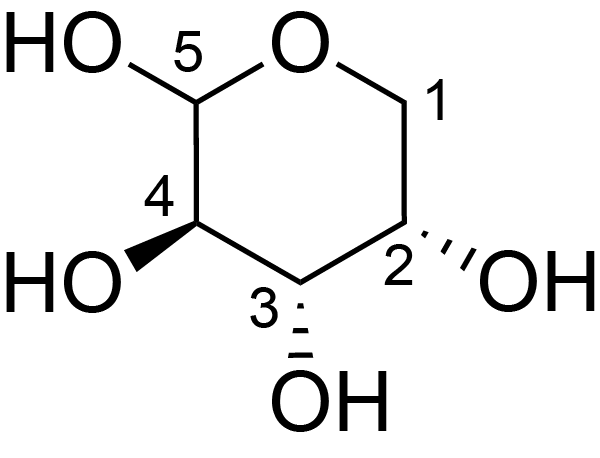
|
H2: 4.2 | 400, 600, and 800 | 298.0 | None | TMS | Removing lipid membranes, cytoplasmic proteins, small molecules, and starch. And being back-exchanged in D2O to reduce the water 1H signal intensities. | Pyae Phyo and Mei Hong et al., Fast MAS 1H-13C correlation NMR for structural investigations of plant cell walls, Journal of Biomolecular NMR, 2019 |
| ccmrd_424 | Rhamnogalacturonan I | (1-4)αDGalpA(1-2)αLRhap | Polysaccharide | plant | Arabidopsis thaliana | a-L-Araf |
|
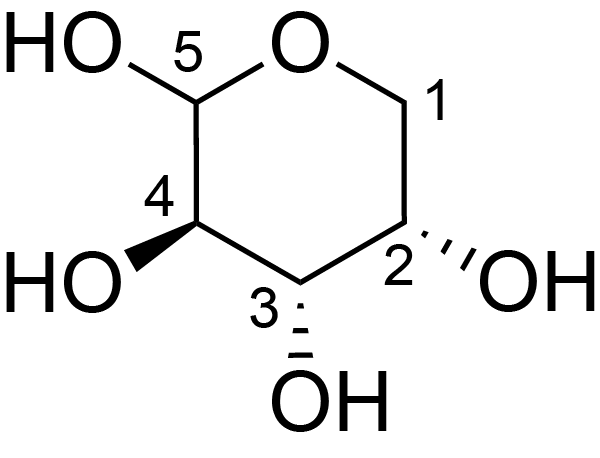
|
H1: 5.3, H2: 4.2 | 400, 600, and 800 | 298.0 | None | TMS | Removing lipid membranes, cytoplasmic proteins, small molecules, and starch. And being back-exchanged in D2O to reduce the water 1H signal intensities. | Pyae Phyo and Mei Hong et al., Fast MAS 1H-13C correlation NMR for structural investigations of plant cell walls, Journal of Biomolecular NMR, 2019 |
| ccmrd_425 | Rhamnogalacturonan I | (1-4)αDGalpA(1-2)αLRhap | Polysaccharide | plant | Arabidopsis thaliana | a-L-Araf |
|
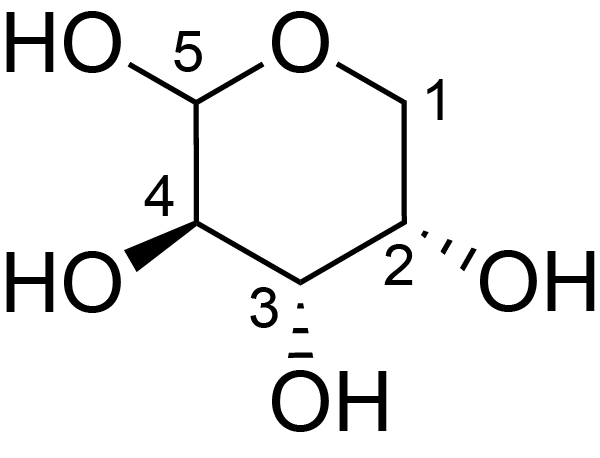
|
H1: 5.1, H2: 4.1, H3: 3.9 | 400, 600, and 800 | 298.0 | None | TMS | Removing lipid membranes, cytoplasmic proteins, small molecules, and starch. And being back-exchanged in D2O to reduce the water 1H signal intensities. | Pyae Phyo and Mei Hong et al., Fast MAS 1H-13C correlation NMR for structural investigations of plant cell walls, Journal of Biomolecular NMR, 2019 |