Complex Carbohydrate Magnetic Resonance Database (CCMRD)
Carbohydrates are crucial for various life activities in living organisms. The Complex Carbohydrates Magnetic Resonance Database (CCMRD), a solid-state NMR database for complex carbohydrates developed at Michigan State University. The Solid-state NMR spectroscopy reveals the molecular structure and dynamics of insoluble complex carbohydrates.
| Database ID | Trival Name | Linear Code | Compound Class | Taxonomy Domain | Species | Residue | SNFG | Chemical Structure | Chemical Shifts (ppm) | Spectrometer (MHz) | Temperature (K) | pH | NMR Reference Compound | Sample Treatment | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ccmrd_406 | Cellulose | Glcp | Polysaccharide | other | Commercial chemical compound | b-D-Glcp |
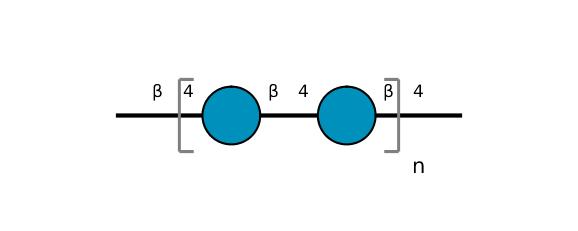
|
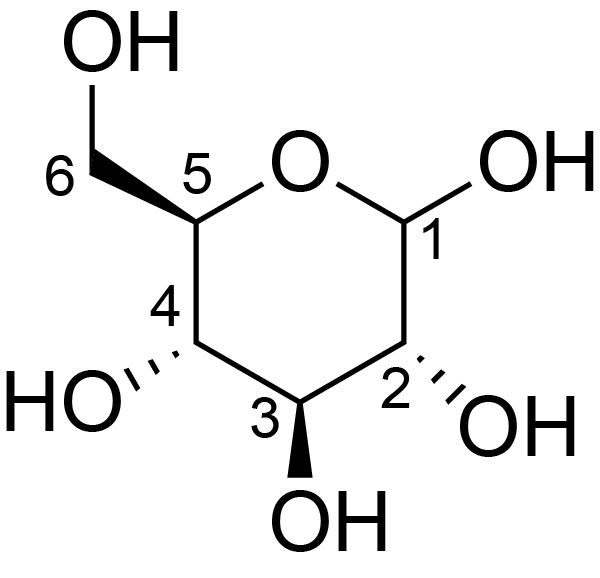
|
C1: 104.0, C2: 74.0, C3: 74.0, C4: 84.0, C5: 74.0, C6: 61.0 | 300 | 298.0 | None | Glycine | Commercial chemical compound | Huiru Tang et al., C MAS NMR studies of the effects of hydration on the cell walls of potatoes and Chinese water chestnuts, Journal of agricultural and food chemistry, 1999 |
| ccmrd_407 | Chitin | GlcpNAc | Polysaccharide | bacteria | Bacillus subtilis | ?-?-GlcpNAc |
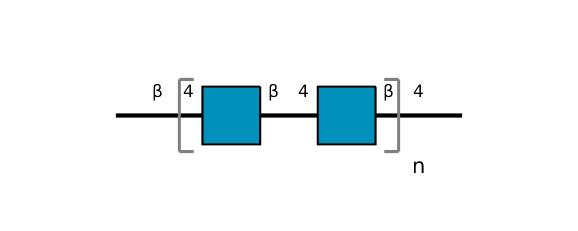
|
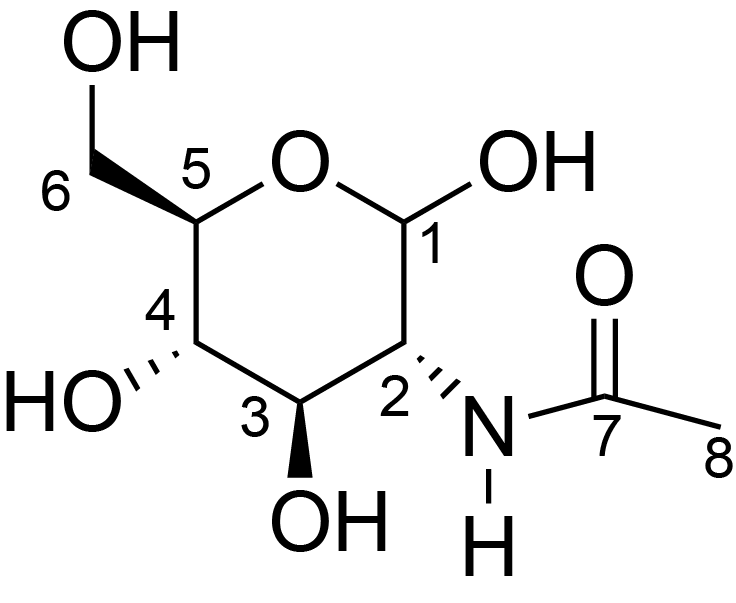
|
C1: 101.0, C2: 56.0, C3: 73.1, C4: 80.4, C5: 75.7, C6: 60.7, C8: 23.0, H1: 4.5, H2: 3.7, H3: 3.6, H4: 3.6, H5: 3.4, H6: 3.7, H8: 2.0, N1: 123.6 | 950 | 273.0 | None | ND | Purified Peptidoglycan | Catherine Bougault et al., Studying intact bacterial peptidoglycan by proton-detected NMR spectroscopy at 100 kHz MAS frequency, Journal of Structural Biology, 2019 |
| ccmrd_408 | N-acetyl muramic acid | MurpNAc | Polysaccharide | bacteria | Bacillus subtilis | ?-?-MurpNAc |
|
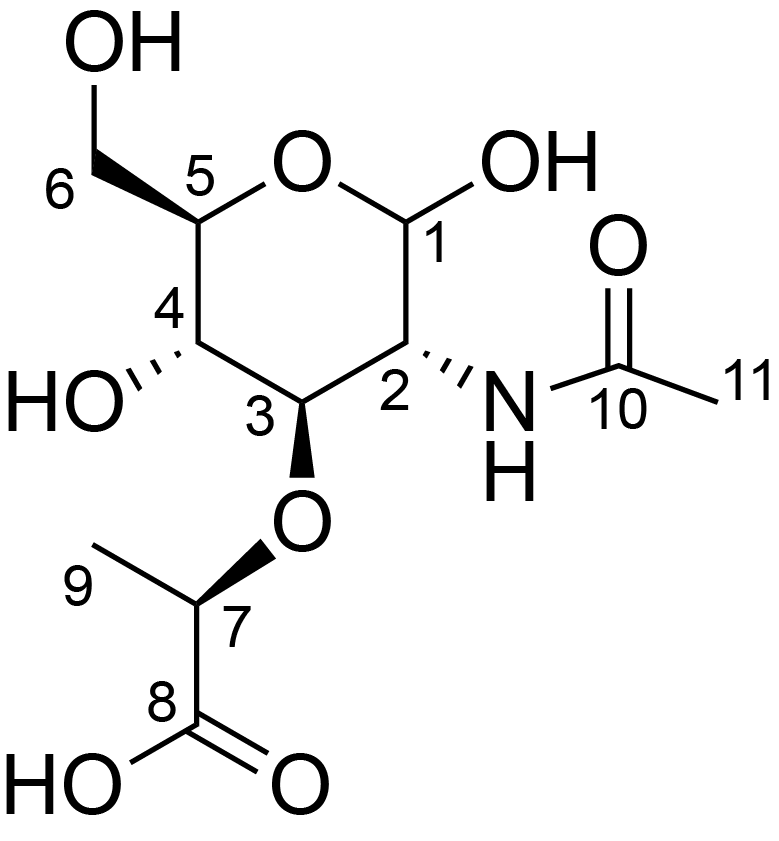
|
C1: 103.6, C2: 56.4, C3: 80.6, C4: 73.7, C5: 76.0, C6: 61.1, C7: 78.6, C9: 18.4, C11: 23.1, H1: 4.4, H2: 3.7, H3: 3.5, H4: 3.3, H5: 3.8, H6: 3.7, H7: 4.4, H9: 1.3, H11: 1.9, N1: 122.6 | 950 | 273.0 | None | ND | Purified Peptidoglycan | Catherine Bougault et al., Studying intact bacterial peptidoglycan by proton-detected NMR spectroscopy at 100 kHz MAS frequency, Journal of Structural Biology, 2019 |
| ccmrd_409 | cellulose | Glcp | Polysaccharide | plant | Picea abies (Norway spruce) | b-D-Glcp |
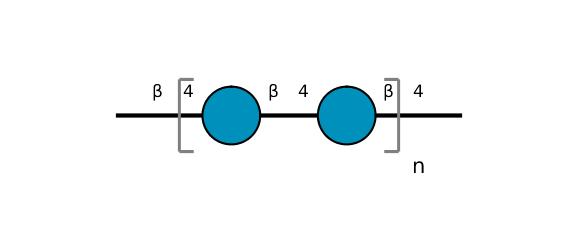
|
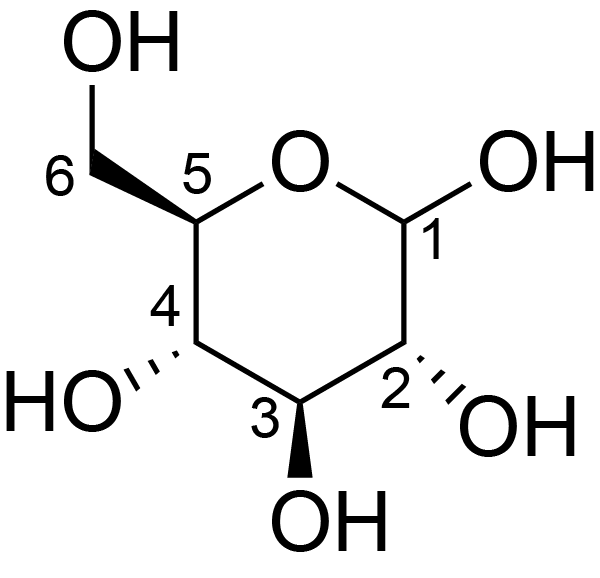
|
C1: 104.3, C4: 88.3, C5: 71.7, C6: 66.0 | 850 | 298.0 | None | TMS | frozen in liquid nitrogen and ground to produce a fine powder | Oliver et al., Molecular, Nature, 2019 |
| ccmrd_410 | cellulose | Glcp | Polysaccharide | plant | Picea abies (Norway spruce) | b-D-Glcp |
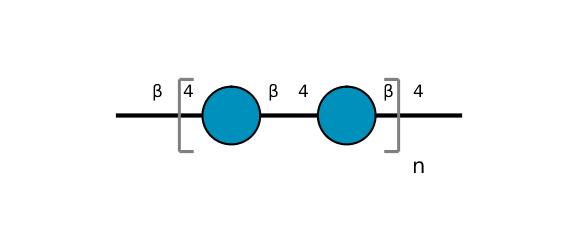
|
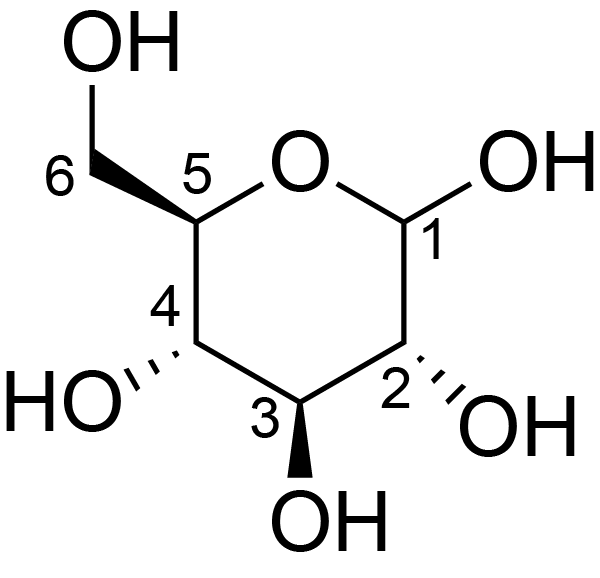
|
C1: 106.0, C2: 71.6, C3: 75.3, C4: 90.0, C5: 71.7, C6: 65.8 | 850 | 298.0 | None | TMS | frozen in liquid nitrogen and ground to produce a fine powder | Oliver M. Terrett et al., Molecular architecture of softwood revealed by solid-state NMR, Nature Communications, 2019 |
| ccmrd_411 | cellulose | Glcp | Polysaccharide | plant | Picea abies (Norway spruce) | b-D-Glcp |
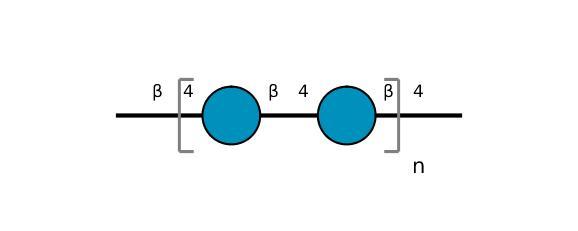
|
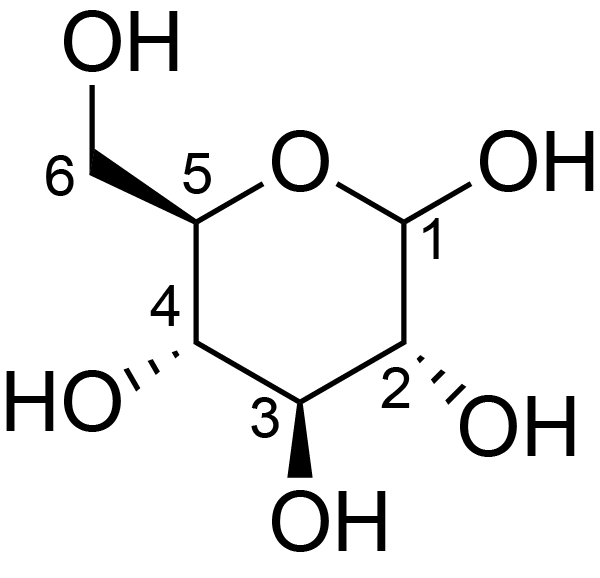
|
C1: 105.4, C2: 72.4, C3: 75.9, C4: 89.5, C5: 72.8, C6: 65.5 | 850 | 298.0 | None | TMS | frozen in liquid nitrogen and ground to produce a fine powder | Oliver M. Terrett et al., Molecular architecture of softwood revealed by solid-state NMR, Nature Communications, 2019 |
| ccmrd_412 | cellulose | Glcp | Polysaccharide | plant | Picea abies (Norway spruce) | b-D-Glcp |
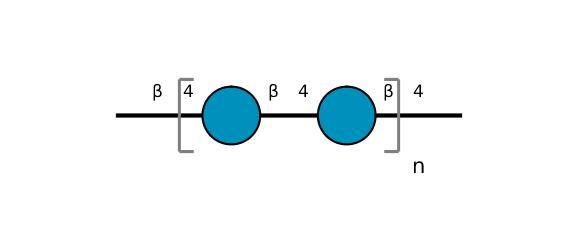
|
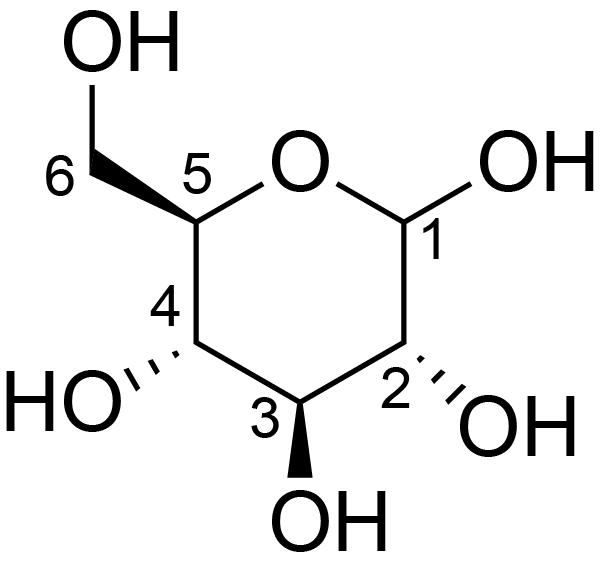
|
C1: 105.3, C2: 72.6, C3: 75.4, C4: 84.1, C5: 74.3, C6: 62.2 | 850 | 298.0 | None | TMS | frozen in liquid nitrogen and ground to produce a fine powder | Oliver M. Terrett et al., Molecular architecture of softwood revealed by solid-state NMR, Nature Communications, 2019 |
| ccmrd_413 | cellulose | Glcp | Polysaccharide | plant | Picea abies (Norway spruce) | b-D-Glcp |
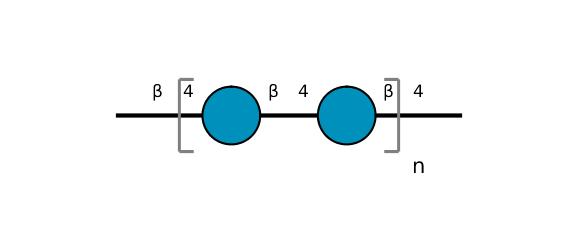
|
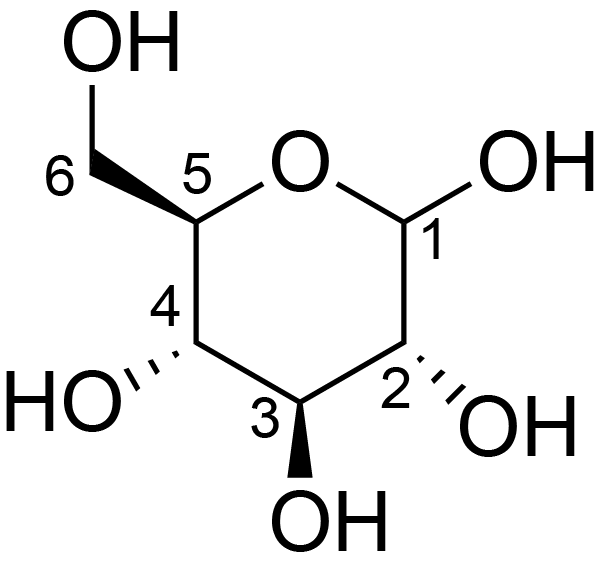
|
C1: 104.3, C4: 88.3, C5: 71.7, C6: 66.0 | 850 | 298.0 | None | TMS | frozen in liquid nitrogen and ground to produce a fine powder | Oliver M. Terrett et al., Molecular architecture of softwood revealed by solid-state NMR, Nature Communications, 2019 |
| ccmrd_414 | cellulose | Glcp | Polysaccharide | plant | Picea abies (Norway spruce) | b-D-Glcp |
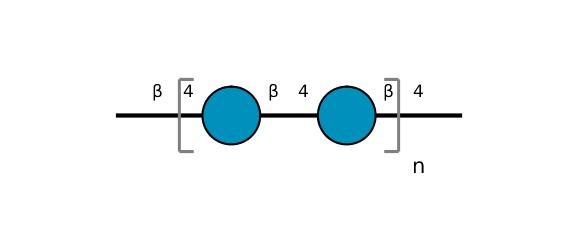
|
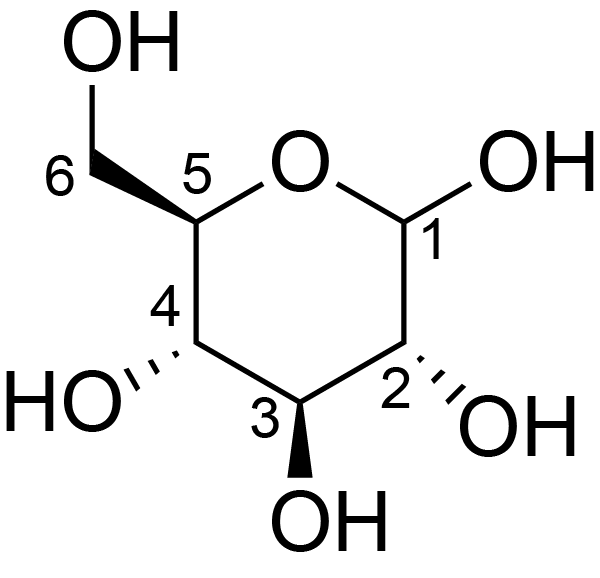
|
C1: 105.2, C3: 74.2, C4: 83.7, C5: 75.6, C6: 61.9 | 850 | 298.0 | None | TMS | frozen in liquid nitrogen and ground to produce a fine powder | Oliver M. Terrett et al., Molecular architecture of softwood revealed by solid-state NMR, Nature Communications, 2019 |
| ccmrd_415 | cellulose | Glcp | Polysaccharide | plant | Picea abies (Norway spruce) | b-D-Glcp |
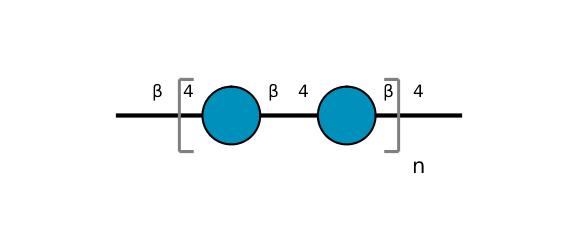
|
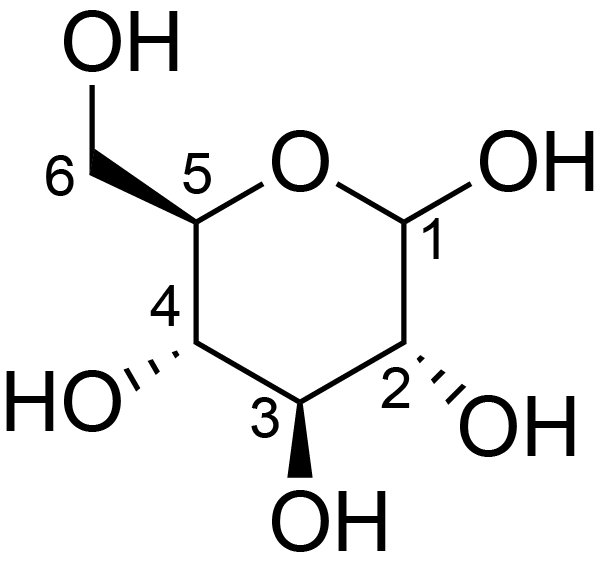
|
C1: 105.3, C2: 72.7, C3: 75.6, C4: 84.5, C5: 75.2, C6: 62.6 | 850 | 298.0 | None | TMS | frozen in liquid nitrogen and ground to produce a fine powder | Oliver M. Terrett et al., Molecular architecture of softwood revealed by solid-state NMR, Nature Communications, 2019 |