Complex Carbohydrate Magnetic Resonance Database (CCMRD)
Carbohydrates are crucial for various life activities in living organisms. The Complex Carbohydrates Magnetic Resonance Database (CCMRD), a solid-state NMR database for complex carbohydrates developed at Michigan State University. The Solid-state NMR spectroscopy reveals the molecular structure and dynamics of insoluble complex carbohydrates.
| Database ID | Trival Name | Linear Code | Compound Class | Taxonomy Domain | Species | Residue | SNFG | Chemical Structure | Chemical Shifts (ppm) | Spectrometer (MHz) | Temperature (K) | pH | NMR Reference Compound | Sample Treatment | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ccmrd_366 | Cellulose | Glcp | Polysaccharide | bacteria | Komagataeibacter xylinus | b-D-Glcp |
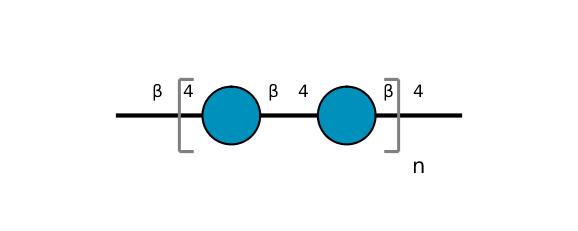
|
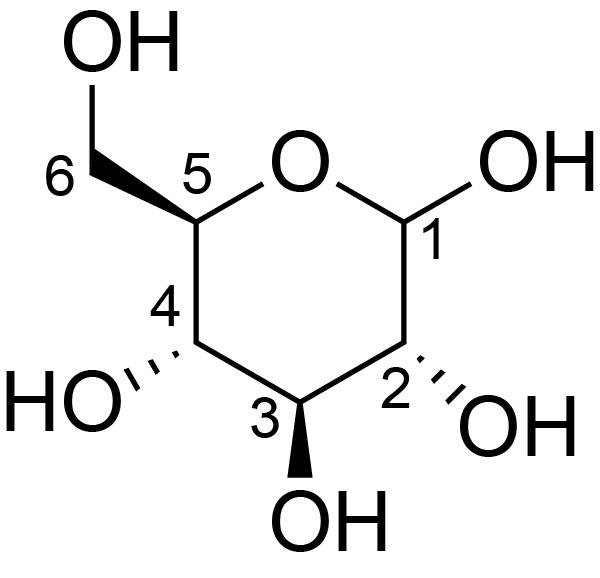
|
C1: 107.2, C2: 72.9, C3: 73.5, C4: 87.7, C5: 75.0, C6: 62.5 | 600 | 298.0 | None | Adamantane | Purified cellulose | Alexander Idström et al., C NMR assignments of regenerated cellulose from solid-state 2D NMR spectroscopy, Carbohydrate Polymers, 2016 |
| ccmrd_367 | Cellulose | Glcp | Polysaccharide | bacteria | Komagataeibacter xylinus | b-D-Glcp |
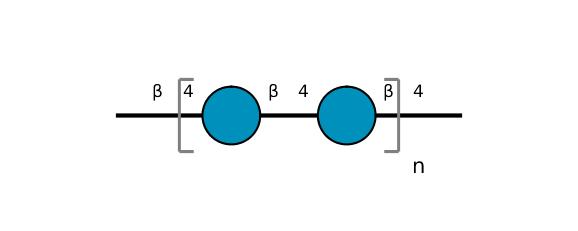
|
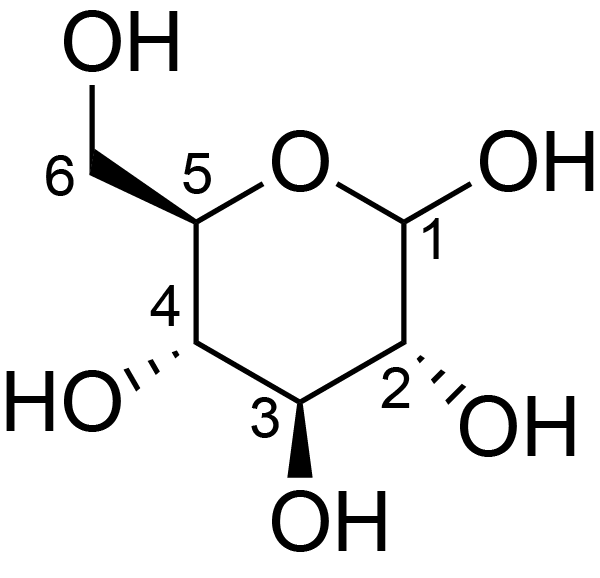
|
C1: 105.1, C2: 74.8, C3: 76.8, C4: 88.9, C5: 72.0, C6: 63.2 | 600 | 298.0 | None | Adamantane | Purified cellulose | Alexander Idström et al., C NMR assignments of regenerated cellulose from solid-state 2D NMR spectroscopy, Carbohydrate Polymers, 2016 |
| ccmrd_368 | Cellulose | Glcp | Polysaccharide | bacteria | Komagataeibacter xylinus | b-D-Glcp |
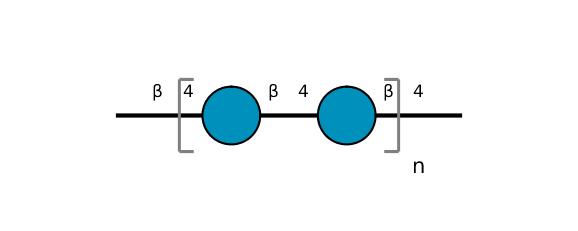
|
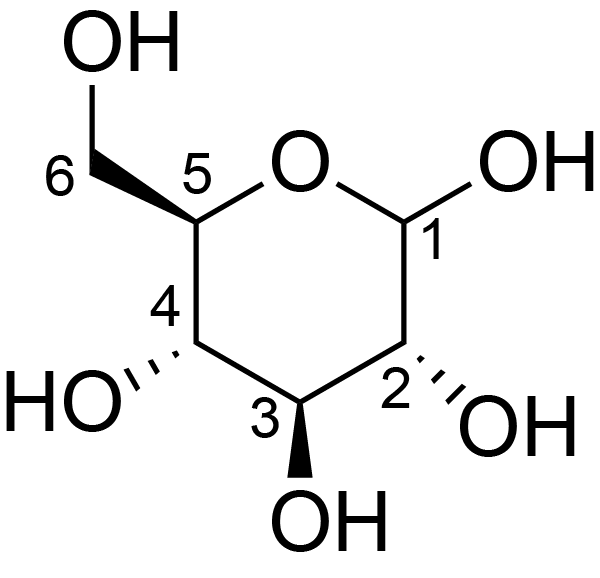
|
C1: 106.4, C2: 72.8, C3: 73.4, C4: 86.2, C5: 75.9, C6: 61.7 | 600 | 298.0 | None | Adamantane | Purified cellulose | Alexander Idström et al., C NMR assignments of regenerated cellulose from solid-state 2D NMR spectroscopy, Carbohydrate Polymers, 2016 |
| ccmrd_369 | Cellulose | Glcp | Polysaccharide | bacteria | Komagataeibacter xylinus | b-D-Glcp |
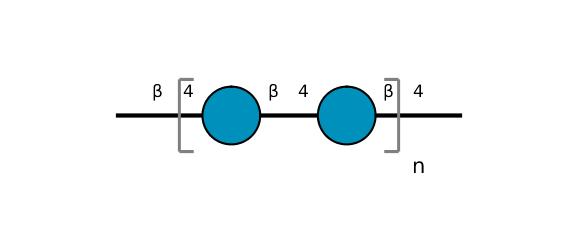
|
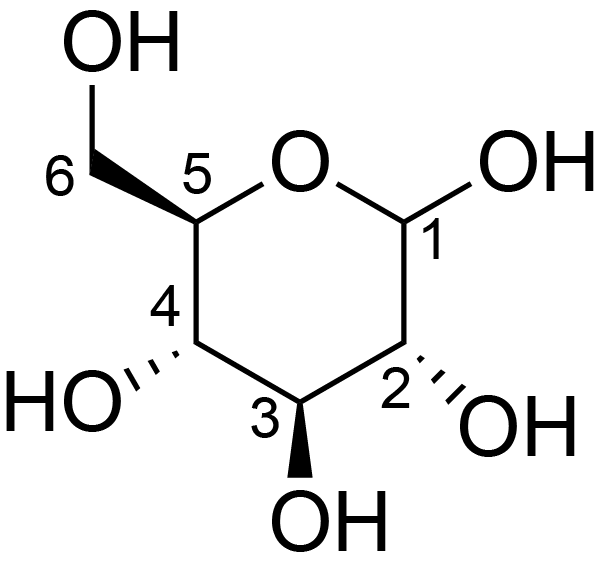
|
C1: 103.3, C2: 73.7, C3: 74.5, C4: 84.8, C5: 74.5, C6: 61.3 | 600 | 298.0 | None | Adamantane | Purified cellulose | Alexander Idström et al., C NMR assignments of regenerated cellulose from solid-state 2D NMR spectroscopy, Carbohydrate Polymers, 2016 |
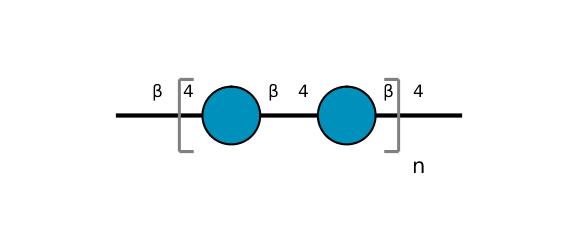
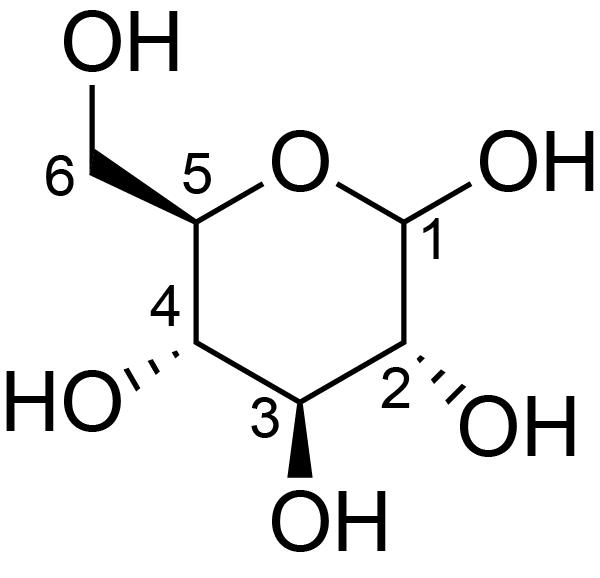
| ccmrd_370 | Cellulose | Glcp | Polysaccharide | plant | Picea abies (L.) H.Karst | b-D-Glcp |
|
|
C1: 105.0, C3: 75.0, C4: 89.0, C5: 75.0, C6: 65.0 | 200 | 298.0 | None | TMS | Ball-milled cellulose | David L. VanderHart et al., Studies of microstructure in native celluloses using solid-state carbon-13 NMR, Macromolecules, 1984 |
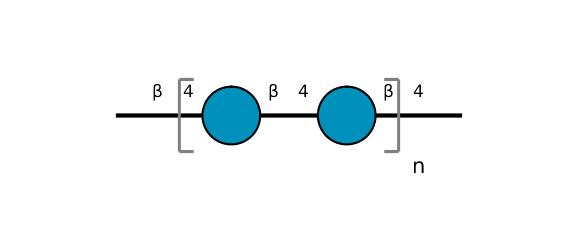
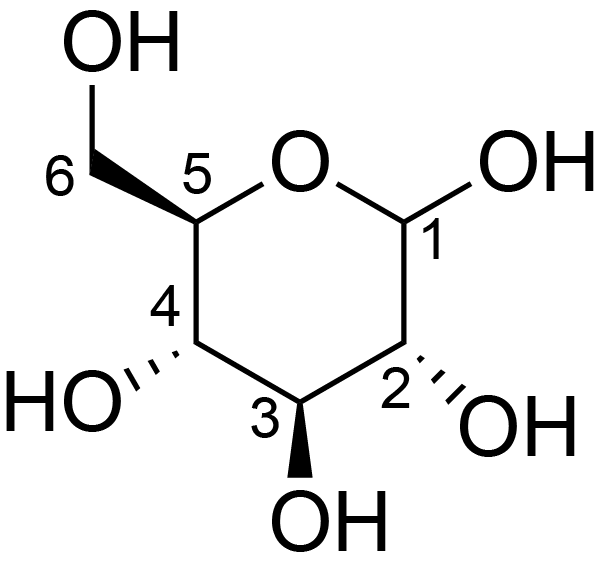
| ccmrd_371 | Cellulose | Glcp | Polysaccharide | plant | Picea abies (L.) H.Karst | b-D-Glcp |
|
|
C1: 105.0, C3: 72.0, C4: 84.0, C6: 61.0 | 200 | 298.0 | None | TMS | Ball-milled cellulose | David L. VanderHart et al., Studies of microstructure in native celluloses using solid-state carbon-13 NMR, Macromolecules, 1984 |
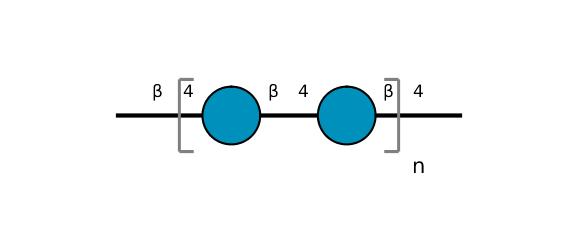
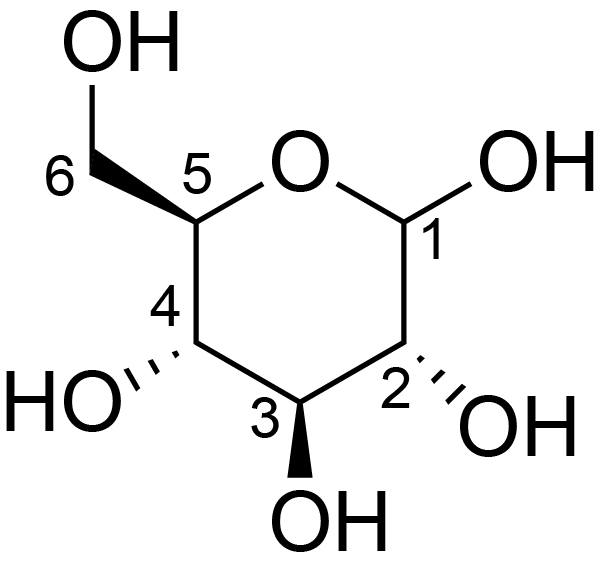
| ccmrd_372 | Cellulose | Glcp | Polysaccharide | plant | wood | b-D-Glcp |
|
|
C1: 106.6, C2: 76.6, C3: 78.3, C4: 90.5, C5: 73.7, C6: 64.7 | 500 | 298.0 | None | ND | Extracted cellulose | Dimitris Sakellariou et al., High-Resolution NMR Correlation Spectra of Disordered Solids, Journal of the American Chemical Society, 2003 |
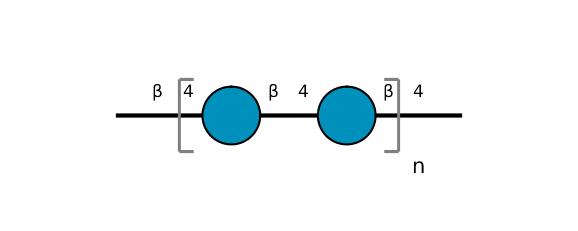
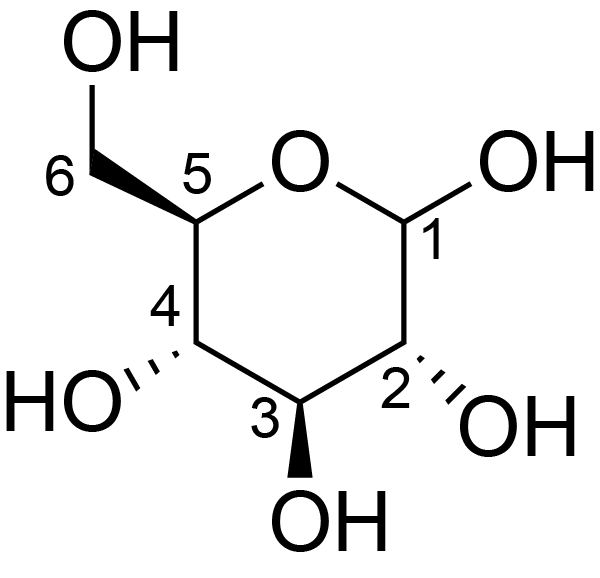
| ccmrd_373 | Cellulose | Glcp | Polysaccharide | plant | wood | b-D-Glcp |
|
|
C1: 108.8, C2: 74.5, C3: 77.8, C4: 89.2, C5: 77.8, C6: 64.1 | 500 | 298.0 | None | ND | Extracted cellulose | Dimitris Sakellariou et al., High-Resolution NMR Correlation Spectra of Disordered Solids, Journal of the American Chemical Society, 2003 |
| ccmrd_374 | Cellulose | Glcp | Polysaccharide | bacteria | Komagataeibacter xylinus | b-D-Glcp |
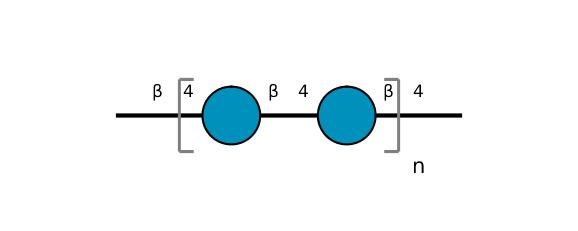
|
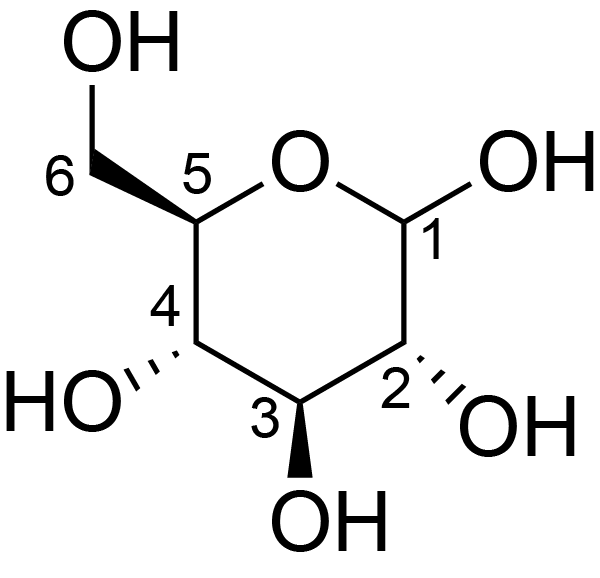
|
C1: 105.0, C2: 71.7, C3: 74.7, C4: 89.8, C5: 70.9, C6: 65.3 | 500 | 298.0 | None | Glycine | Purified cellulose | Keiko Okushita et al., Statistical approach for solid-state NMR spectra of cellulose derived from a series of variable parameters, Polymer journal, 2012 |
| ccmrd_375 | Cellulose | Glcp | Polysaccharide | bacteria | Komagataeibacter xylinus | b-D-Glcp |
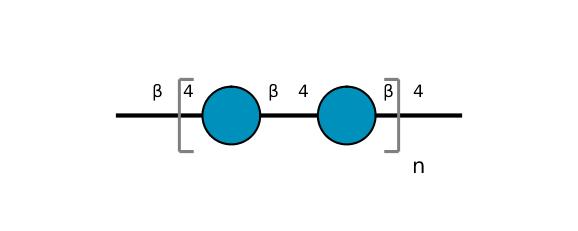
|
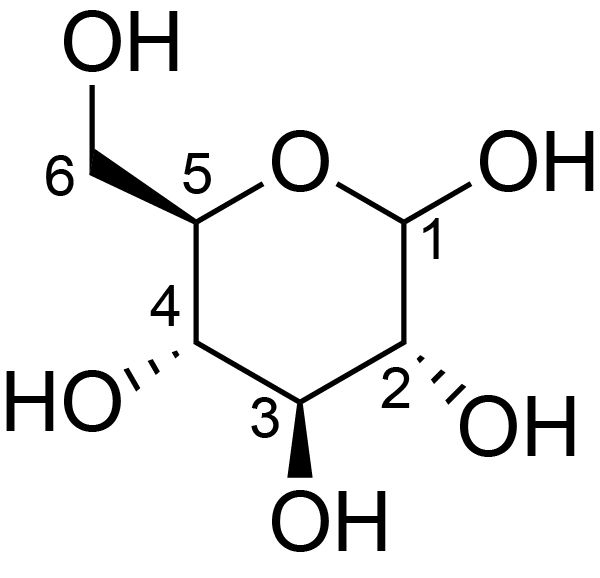
|
C1: 103.9, C2: 70.9, C4: 88.9, C5: 72.6 | 500 | 298.0 | None | Glycine | Purified cellulose | Keiko Okushita et al., Statistical approach for solid-state NMR spectra of cellulose derived from a series of variable parameters, Polymer journal, 2012 |