Complex Carbohydrate Magnetic Resonance Database (CCMRD)
Carbohydrates are crucial for various life activities in living organisms. The Complex Carbohydrates Magnetic Resonance Database (CCMRD), a solid-state NMR database for complex carbohydrates developed at Michigan State University. The Solid-state NMR spectroscopy reveals the molecular structure and dynamics of insoluble complex carbohydrates.
| Database ID | Trival Name | Linear Code | Compound Class | Taxonomy Domain | Species | Residue | SNFG | Chemical Structure | Chemical Shifts (ppm) | Spectrometer (MHz) | Temperature (K) | pH | NMR Reference Compound | Sample Treatment | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ccmrd_325 | N-acetyl muramic acid | MurpNAc | Peptidoglycan | bacteria | Escherichia coli K12 | b-?-MurpNAc |
|
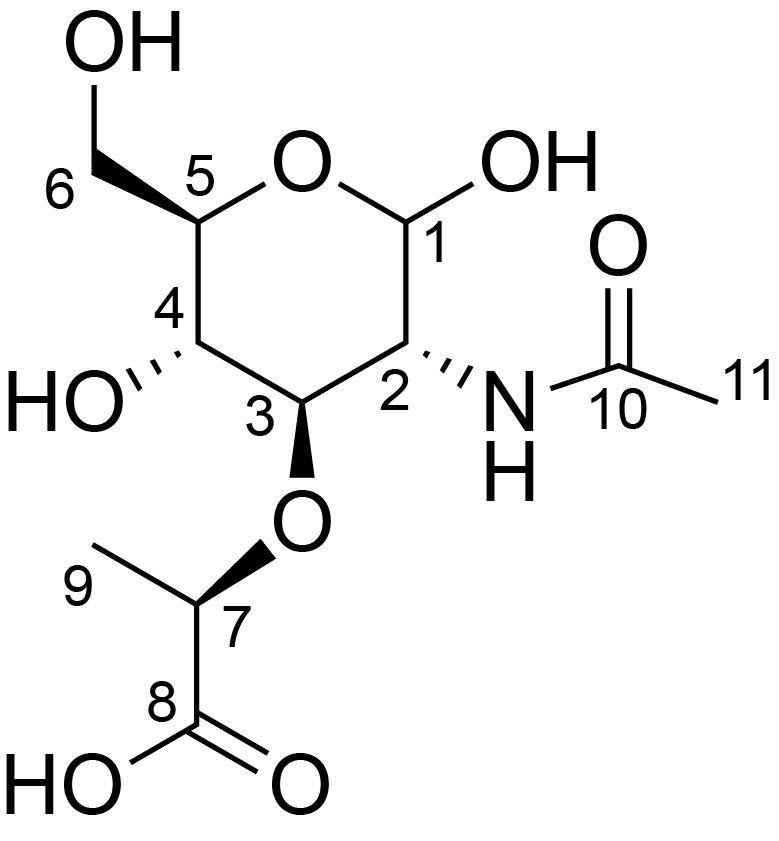
|
C1: 101.1, C2: 56.8, C3: 73.6 | 600 | 298.0 | None | ND | Lyophylized LPS | Cedric Laguri et al., Solid State NMR Studies of Intact Lipopolysaccharide Endotoxin, ACS chemical biology, 2018 |
| ccmrd_326 | Keto-deoxyoctulosonate | Kdop | Lipopolysaccharide | bacteria | Pseudomonas aeruginosa | a-?-Kdop |
|
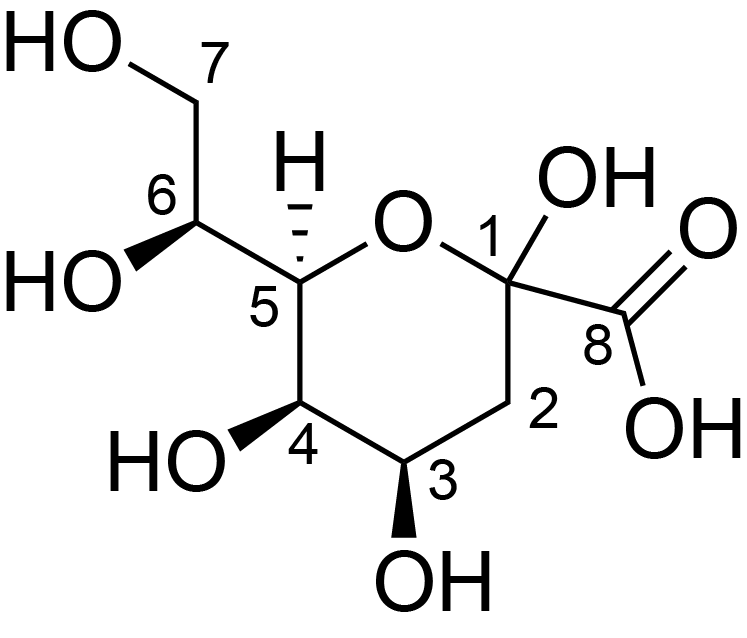
|
C1: 176.3, C3: 35.4, C4: 73.1 | 600 | 298.0 | 6.0 | ND | Lyophylized LPS | Cedric Laguri et al., Solid State NMR Studies of Intact Lipopolysaccharide Endotoxin, ACS chemical biology, 2018 |
| ccmrd_327 | Heptose | Hepp | Lipopolysaccharide | bacteria | Pseudomonas aeruginosa | a-?-Hepp |
|
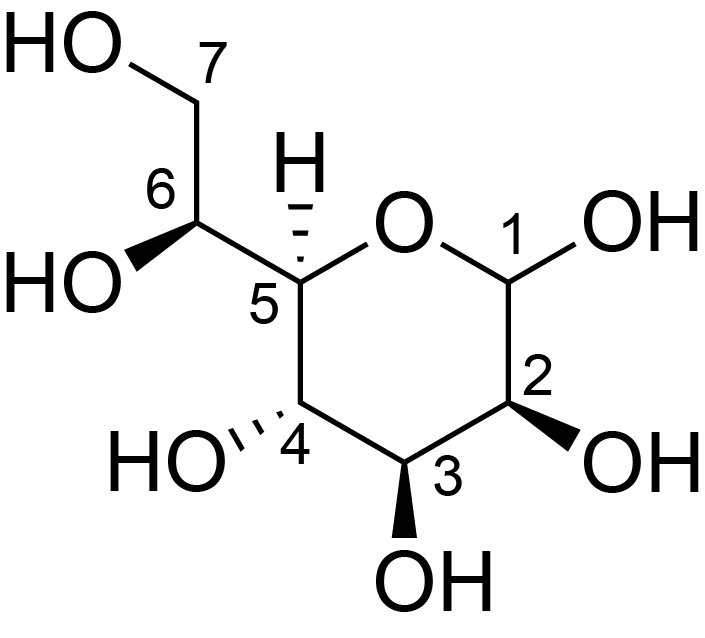
|
C1: 105.3, C2: 74.6, C3: 76.8, C4: 70.6, C5: 77.0, C7: 61.8 | 600 | 298.0 | 6.0 | ND | Lyophylized LPS | Cedric Laguri et al., Solid State NMR Studies of Intact Lipopolysaccharide Endotoxin, ACS chemical biology, 2018 |
| ccmrd_328 | Rhamnose | Rhap | Lipopolysaccharide | bacteria | Pseudomonas aeruginosa | a-?-Rhap |

|
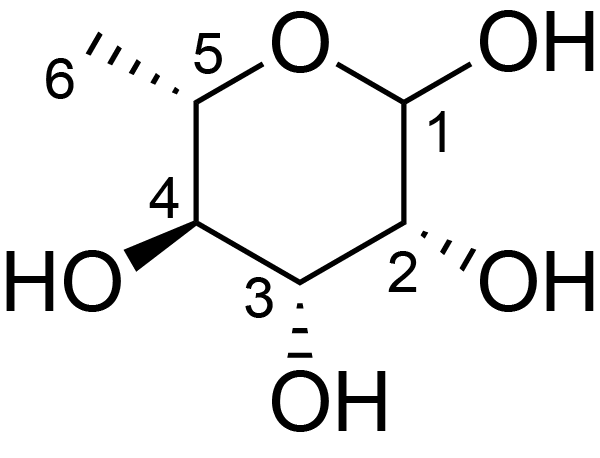
|
C1: 101.2, C2: 71.2, C3: 81.2, C4: 73.8, C5: 70.2, C6: 18.1 | 600 | 298.0 | 6.0 | ND | Lyophylized LPS | Cedric Laguri et al., Solid State NMR Studies of Intact Lipopolysaccharide Endotoxin, ACS chemical biology, 2018 |
| ccmrd_329 | Rhamnan | Rhap | Lipopolysaccharide | bacteria | Pseudomonas aeruginosa | a-?-Rhap |
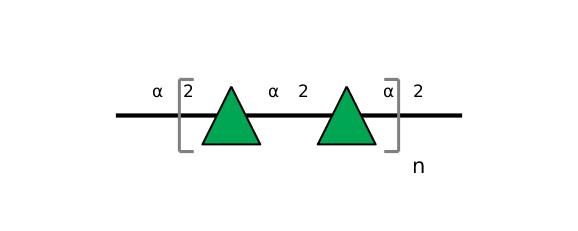
|
|
C1: 101.9, C2: 79.3, C3: 73.7, C4: 71.1, C5: 70.4, C6: 17.8, H1: 5.2, H2: 4.1, H3: 3.6, H5: 3.9, H6: 1.3 | 600 | 298.0 | 6.0 | ND | Lyophylized LPS | Cedric Laguri et al., Solid State NMR Studies of Intact Lipopolysaccharide Endotoxin, ACS chemical biology, 2018 |
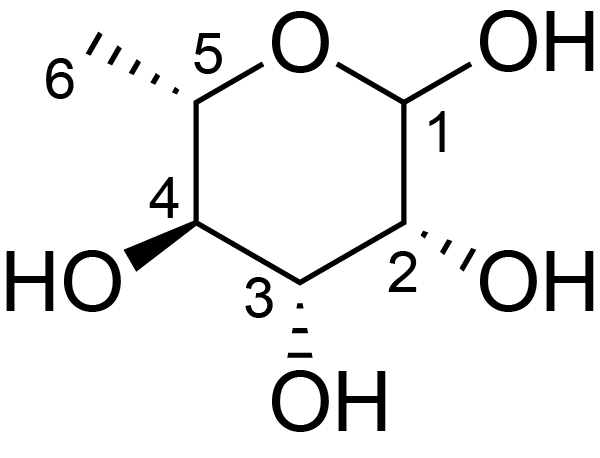
| ccmrd_330 | Rhamnan | Rhap | Lipopolysaccharide | bacteria | Pseudomonas aeruginosa | a-?-Rhap |
|
|
C1: 103.3, C2: 71.1, H1: 5.0, H2: 4.2 | 600 | 298.0 | 6.0 | ND | Lyophylized LPS | Cedric Laguri et al., Solid State NMR Studies of Intact Lipopolysaccharide Endotoxin, ACS chemical biology, 2018 |
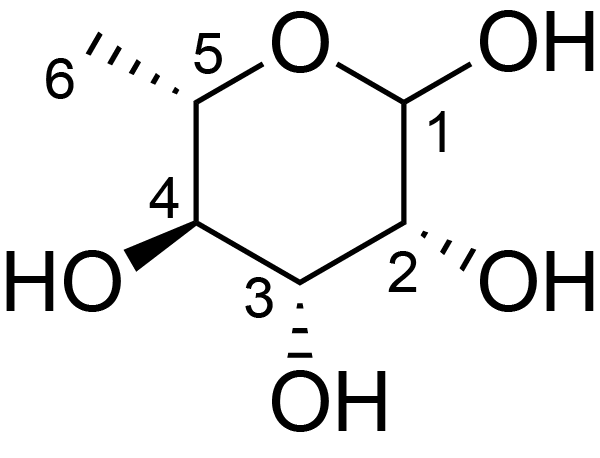
| ccmrd_331 | Rhamnan | Rhap | Lipopolysaccharide | bacteria | Pseudomonas aeruginosa | a-?-Rhap |
|
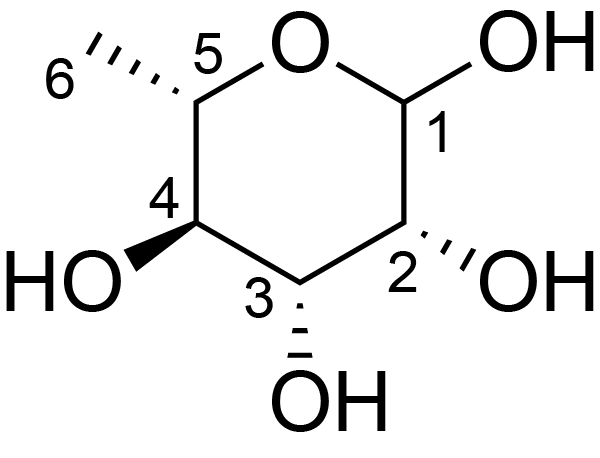
|
C1: 102.5, C2: 67.4, H1: 5.1, H2: 4.2 | 600 | 298.0 | 6.0 | ND | Lyophylized LPS | Cedric Laguri et al., Solid State NMR Studies of Intact Lipopolysaccharide Endotoxin, ACS chemical biology, 2018 |
| ccmrd_332 | Rhamnan | Rhap | Lipopolysaccharide | bacteria | Pseudomonas aeruginosa | ?-?-Rhap |
|
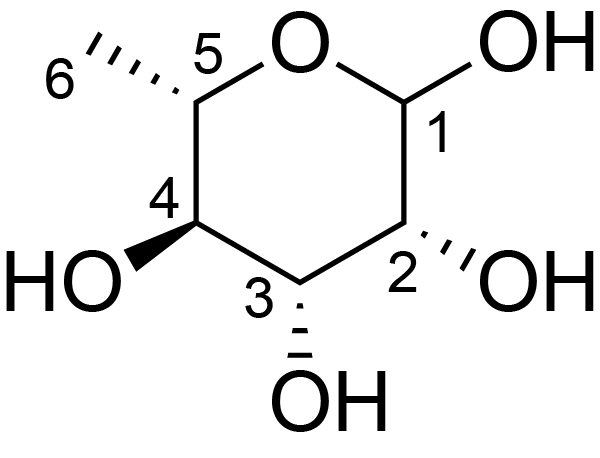
|
H4: 3.5 | 600 | 298.0 | 6.0 | ND | Lyophylized LPS | Cedric Laguri et al., Solid State NMR Studies of Intact Lipopolysaccharide Endotoxin, ACS chemical biology, 2018 |
| ccmrd_333 | Fucose | Fucp | Lipopolysaccharide | bacteria | Pseudomonas aeruginosa | a-?-Fucp |

|
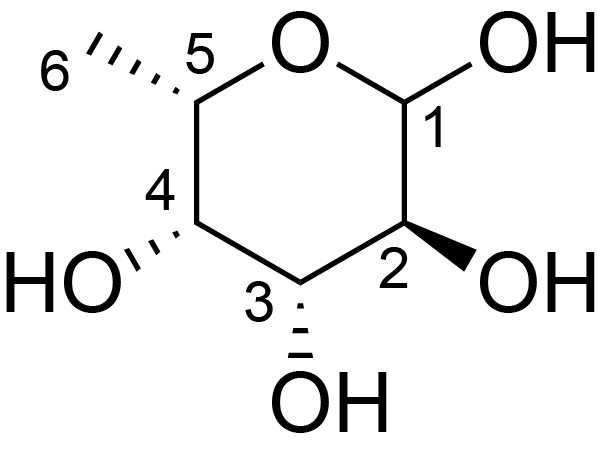
|
C1: 98.4, C2: 48.7, C3: 79.1, C4: 70.9, C5: 68.1, C6: 16.5, H1: 4.9, H2: 4.1, H3: 3.9, H4: 4.0, H5: 4.0, H6: 1.1 | 600 | 298.0 | 6.0 | ND | Lyophylized LPS | Cedric Laguri et al., Solid State NMR Studies of Intact Lipopolysaccharide Endotoxin, ACS chemical biology, 2018 |
| ccmrd_334 | N-acylated mannuronic acids | ManpNAc | Lipopolysaccharide | bacteria | Pseudomonas aeruginosa | b-?-ManpNAc |
|
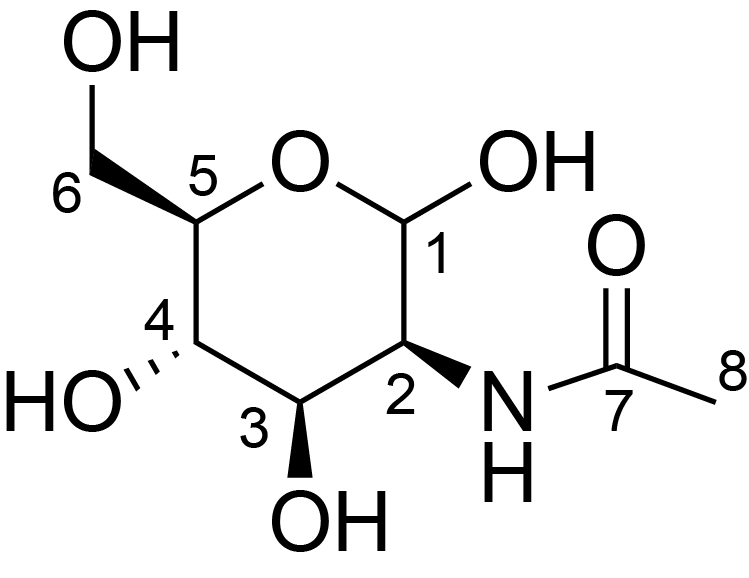
|
C1: 100.9, C2: 52.7, C3: 52.8, C4: 77.0, C5: 78.2, C6: 176.4, H1: 4.9, H2: 4.3, H3: 4.1, H4: 3.7, H5: 3.8 | 600 | 298.0 | 6.0 | ND | Lyophylized LPS | Cedric Laguri et al., Solid State NMR Studies of Intact Lipopolysaccharide Endotoxin, ACS chemical biology, 2018 |