Complex Carbohydrate Magnetic Resonance Database (CCMRD)
Carbohydrates are crucial for various life activities in living organisms. The Complex Carbohydrates Magnetic Resonance Database (CCMRD), a solid-state NMR database for complex carbohydrates developed at Michigan State University. The Solid-state NMR spectroscopy reveals the molecular structure and dynamics of insoluble complex carbohydrates.
| Database ID | Trival Name | Linear Code | Compound Class | Taxonomy Domain | Species | Residue | SNFG | Chemical Structure | Chemical Shifts (ppm) | Spectrometer (MHz) | Temperature (K) | pH | NMR Reference Compound | Sample Treatment | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ccmrd_151 | Rhamnogalacturonan | (1-4)αDGalpA(1-2)αLRhap | Polysaccharide | plant | Arabidopsis thaliana | a-D-GalpA |
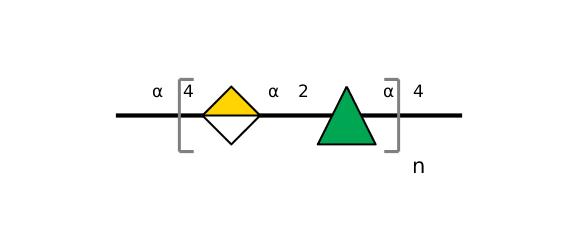
|
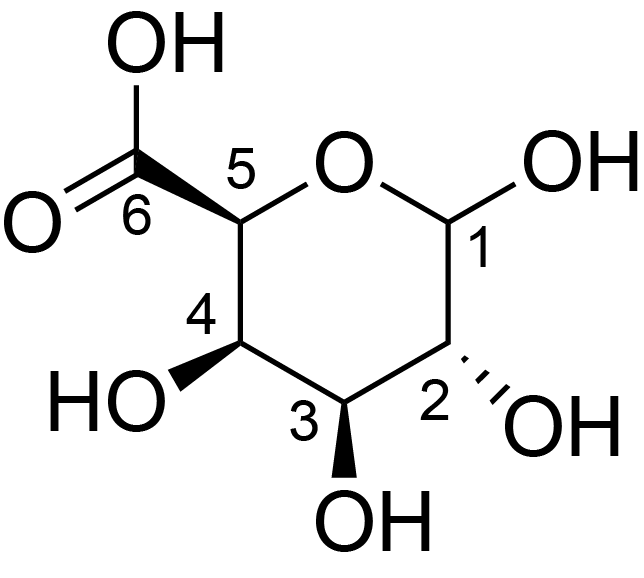
|
C1: 100.1, C2: 68.9, C3: 71.1 | 800 | 298.0 | None | TMS | Whole cell | Pyae Phyo et al., Gradients in Wall Mechanics and Polysaccharides along Growing Inflorescence Stems, Plant physiology, 2017 |
| ccmrd_152 | Rhamnogalacturonan | (1-4)αDGalpA(1-2)αLRhap | Polysaccharide | plant | Arabidopsis thaliana | a-D-GalpA |
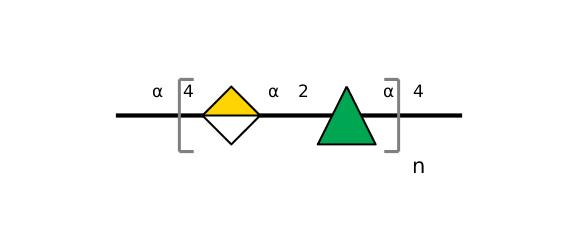
|
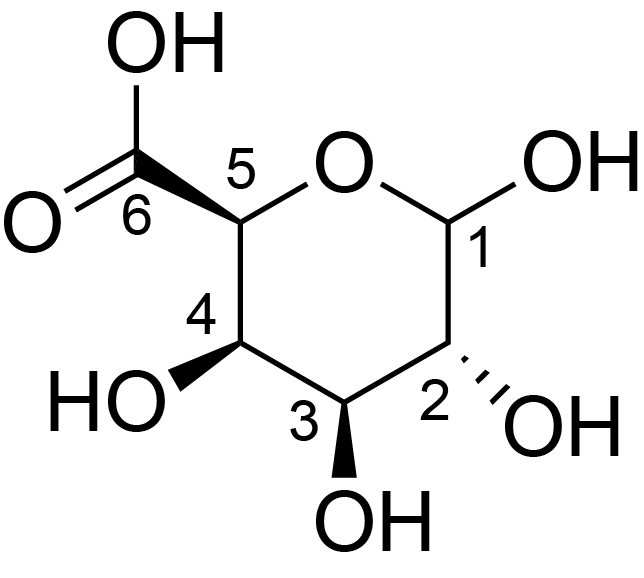
|
C1: 99.1, C2: 67.9 | 800 | 298.0 | None | TMS | Whole cell | Pyae Phyo et al., Gradients in Wall Mechanics and Polysaccharides along Growing Inflorescence Stems, Plant physiology, 2017 |
| ccmrd_153 | Rhamnogalacturonan | (1-4)αDGalpA(1-2)αLRhap | Polysaccharide | plant | Arabidopsis thaliana | a-D-GalpA |
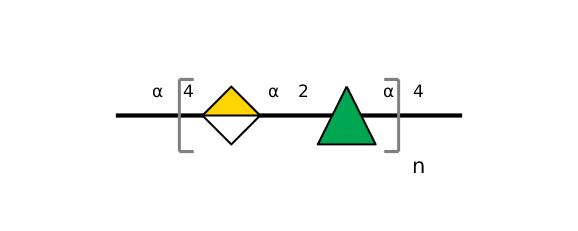
|
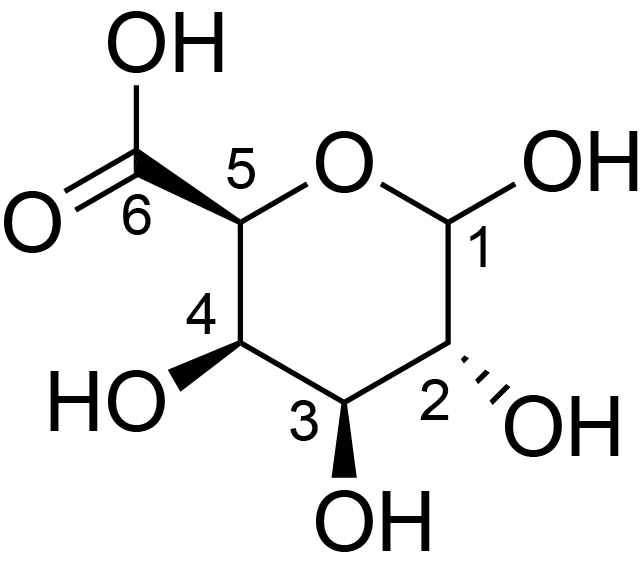
|
C1: 99.0, C2: 68.6 | 800 | 298.0 | None | TMS | Whole cell | Pyae Phyo et al., Gradients in Wall Mechanics and Polysaccharides along Growing Inflorescence Stems, Plant physiology, 2017 |
| ccmrd_154 | Rhamnogalacturonan | (1-4)αDGalpA(1-2)αLRhap | Polysaccharide | plant | Arabidopsis thaliana | a-D-GalpA |
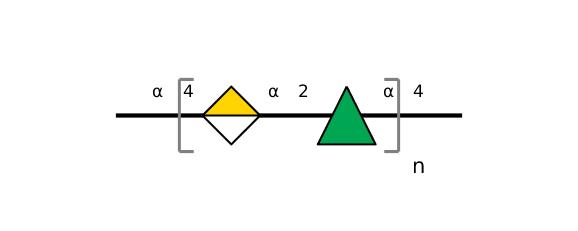
|
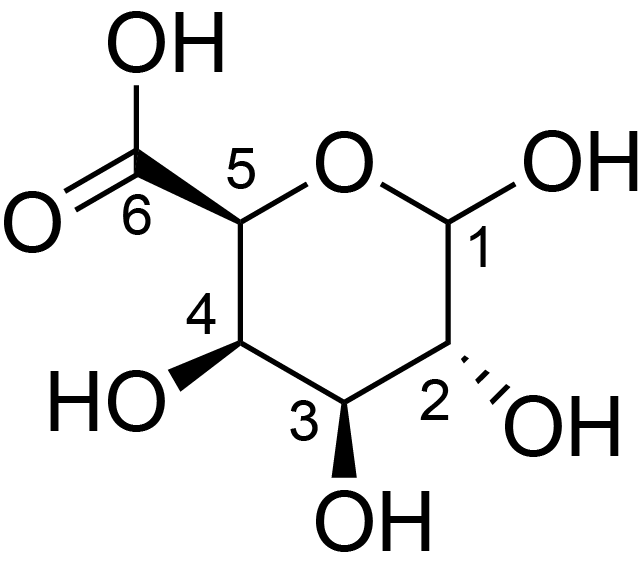
|
C1: 98.4, C2: 68.8 | 800 | 298.0 | None | TMS | Whole cell | Pyae Phyo et al., Gradients in Wall Mechanics and Polysaccharides along Growing Inflorescence Stems, Plant physiology, 2017 |
| ccmrd_155 | Cellulose | Glcp | Polysaccharide | plant | Linum usitatissimum | b-D-Glcp |
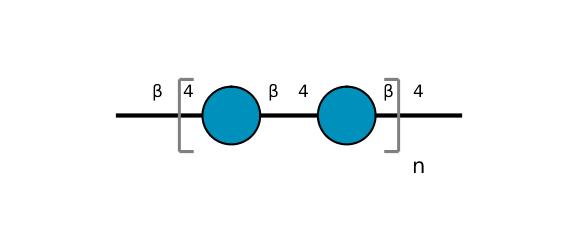
|
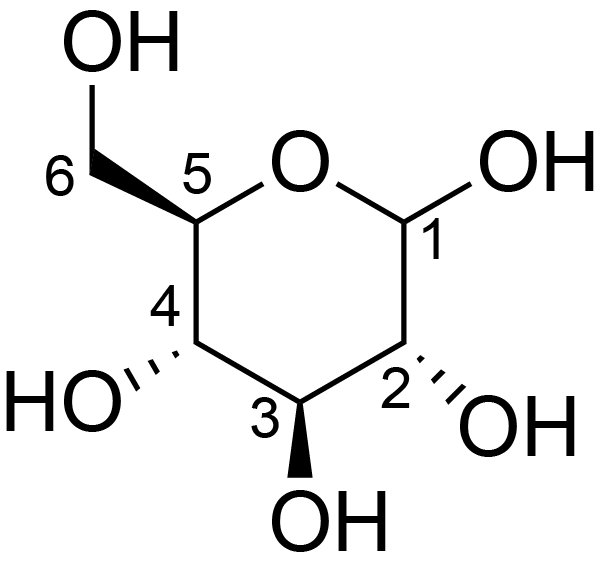
|
C1: 105.2, C2: 72.3, C3: 73.9, C4: 84.0, C5: 75.5 | 300 | 298.0 | None | ND | Acid hydrolysis and extraction cell | Remco J. Vietor et al., Conformational features of crystal-surface cellulose from higher plants, The Plant journal, 2002 |
| ccmrd_156 | Cellulose | Glcp | Polysaccharide | plant | Linum usitatissimum | b-D-Glcp |
|
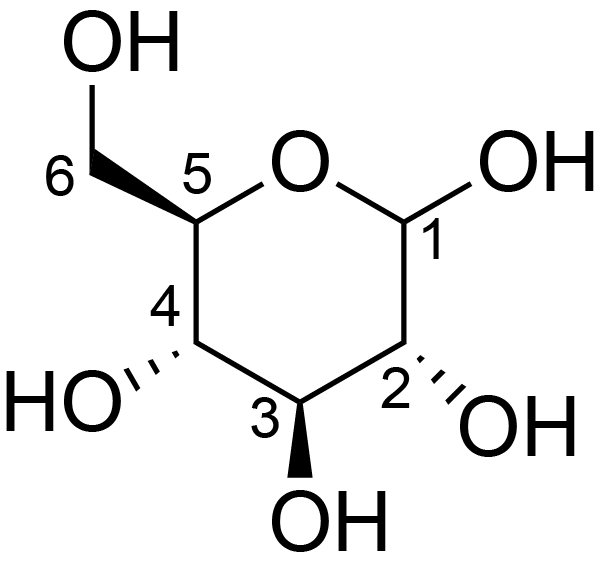
|
C6: 65.5 | 300 | 298.0 | None | ND | Acid hydrolysis and extraction cell | Remco J. Vietor et al., Conformational features of crystal-surface cellulose from higher plants, The Plant journal, 2002 |
| ccmrd_157 | Cellulose | Glcp | Polysaccharide | plant | Linum usitatissimum | b-D-Glcp |
|
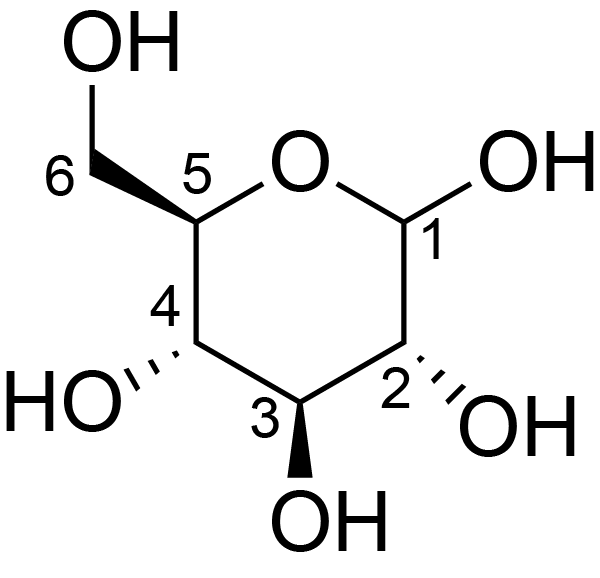
|
C6: 62.8 | 300 | 298.0 | None | ND | Acid hydrolysis and extraction cell | Remco J. Vietor et al., Conformational features of crystal-surface cellulose from higher plants, The Plant journal, 2002 |
| ccmrd_158 | Cellulose | Glcp | Polysaccharide | plant | Linum usitatissimum | b-D-Glcp |
|
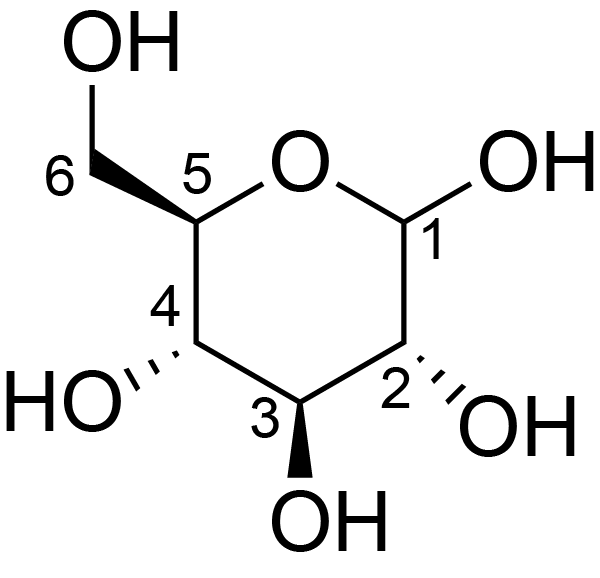
|
C6: 61.5 | 300 | 298.0 | None | ND | Acid hydrolysis and extraction cell | Remco J. Vietor et al., Conformational features of crystal-surface cellulose from higher plants, The Plant journal, 2002 |
| ccmrd_159 | Cellulose | Glcp | Polysaccharide | plant | Apium graveolens | b-D-Glcp |
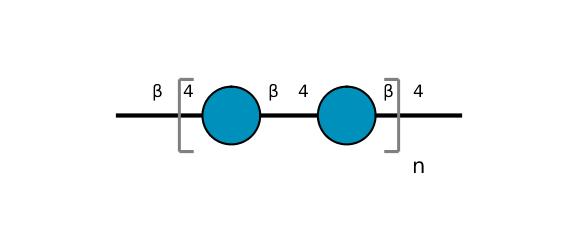
|
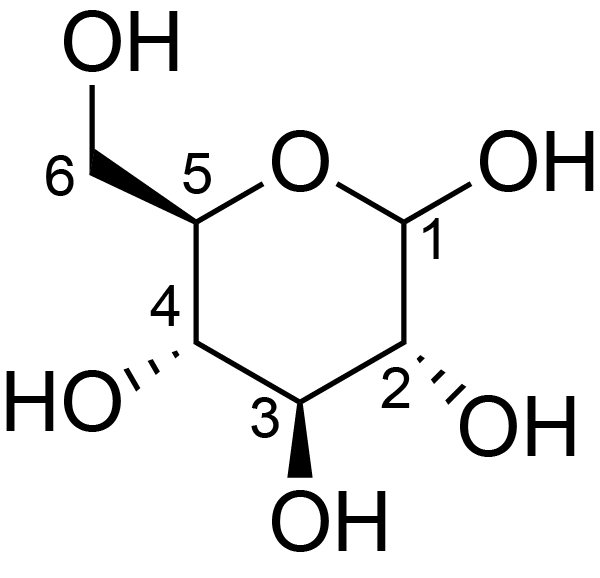
|
C1: 105.0, C2: 72.8, C3: 74.4, C4: 84.0, C5: 75.7, C6: 65.0 | 300 | 298.0 | None | ND | Acid hydrolysis and extraction cell | Remco J. Vietor et al., Conformational features of crystal-surface cellulose from higher plants, The Plant journal, 2002 |
| ccmrd_160 | Cellulose | Glcp | Polysaccharide | plant | Apium graveolens | b-D-Glcp |
|
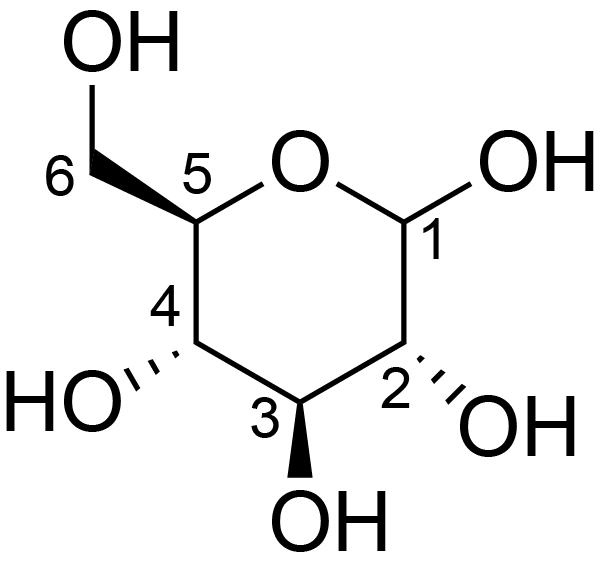
|
C6: 62.8 | 300 | 298.0 | None | ND | Acid hydrolysis and extraction cell | Remco J. Vietor et al., Conformational features of crystal-surface cellulose from higher plants, The Plant journal, 2002 |